Abstract
Microbial metabolisms and interactions that facilitate subsurface conversions of recalcitrant carbon to methane are poorly understood. We deployed an in situ enrichment device in a subsurface coal seam in the Powder River Basin (PRB), USA, and used BONCAT-FACS-Metagenomics to identify translationally active populations involved in methane generation from a variety of coal-derived aromatic hydrocarbons. From the active fraction, high-quality metagenome-assembled genomes (MAGs) were recovered for the acetoclastic methanogen, Methanothrix paradoxum, and a novel member of the Chlorobi with the potential to generate acetate via the Pta-Ack pathway. Members of the Bacteroides and Geobacter also encoded Pta-Ack and together, all four populations had the putative ability to degrade ethylbenzene, phenylphosphate, phenylethanol, toluene, xylene, and phenol. Metabolic reconstructions, gene analyses, and environmental parameters also indicated that redox fluctuations likely promote facultative energy metabolisms in the coal seam. The active “Chlorobi PRB” MAG encoded enzymes for fermentation, nitrate reduction, and multiple oxygenases with varying binding affinities for oxygen. “M. paradoxum PRB” encoded an extradiol dioxygenase for aerobic phenylacetate degradation, which was also present in previously published Methanothrix genomes. These observations outline underlying processes for bio-methane from subbituminous coal by translationally active populations and demonstrate activity-based metagenomics as a powerful strategy in next generation physiology to understand ecologically relevant microbial populations.
Similar content being viewed by others
Introduction
Methane is an important source of energy globally, and in recent years has undergone rapid development accounting for almost 40% of energy consumption in the United States [1]. As energy demands increase and natural gas resources are depleted, there is a heightened need for alternative and less conventional energy technologies to be developed [1]. One near-term, unconventional energy resource is biogenic coalbed methane (CBM), i.e., the biological conversion of coal to methane. It has been estimated that roughly 40% of CBM in the United States is of biogenic origin, and the interest in CBM is growing due to the presence of this natural process associated with many coal reserves in the United States [2]. Biogenic CBM is a source of cleaner energy compared to coal owing to its naturally refined low molecular weight hydrocarbon content and cleaner burning properties [3, 4]. However, methane has 84 times the global warming potential of carbon dioxide over a 20-year period [5], and methane off-gassing at oil and coal wells is both a major safety concern and a serious environmental problem. Ultimately, many hydrocarbon environments can be associated with biogenic methane, and whether the goal is to stimulate methane production for harvesting cleaner fuels or mitigate methane production to restrict carbon release, current understanding indicates the likely rate-limiting step is conversion of the coal to methanogenic precursors [6, 7]. The significance of different carbon cycling pathways involved in the turnover of recalcitrant carbon to methane is still a topic of debate, and unknown carbon cycling pathways continue to be discovered. This fundamental knowledge is necessary to understand microbial processes that contribute to subsurface carbon turnover in relationship to biogenic methane and helps identify unknown pathways that link terrestrial subsurface carbon cycling with carbon dioxide and methane.
Broadly, biogenic CBM can be divided into two primary microbial steps: (i) conversion of coal or other complex organic intermediates into simpler organic precursors and (ii) conversion of simple organic intermediates to methane by methanogens. Coal is a complex heterogeneous hydrocarbon with mixed chemical composition including high-molecular-weight polycyclic aromatic hydrocarbons and derivatives with a high mass fraction of carbon [8]. Coal is classified by ranking thermal maturity with designations increasing from lignite, subbituminous, bituminous, and anthracite. Lower rank subbituminous coals such as those found in the Powder River Basin (PRB, southeastern Montana/northeastern Wyoming, USA) are thought to be more bioavailable than higher rank coals because lower rank coals contain more oxygen, sulfur, and nitrogen and less aromaticity [9]. Laboratory studies have indicated microbial production of CBM can also increase porosity and consequently the bioavailability of coal through the utilization of oxygen-containing functional groups which reduces the degree of crystallization and increases the pore connectivity [10, 11]. Other researchers have shown that low ranking subbituminous coal can have higher concentrations of extractable acetate [12], an important intermediate in microbial conversions of coal to methane [13, 14]. Microbial communities break down coal components into simple intermediates that can be utilized by methanogens to produce methane, but the specific components of the coal that are targeted for degradation and the responsible microbial populations remain unknown.
During methanogenesis methane gas is produced as the final step of organic matter degradation in anoxic environments by taxonomically diverse archaea including seven orders of Euryarchaeota [15, 16], Verstraetearchaeota [17], and possibly Korarchaeota [18, 19]. The primary substrates for archaeal methanogenesis include carbon dioxide and hydrogen, methylated compounds, and acetate [20]. Acetoclastic methanogenesis is thought to be predominant in nature, with estimates suggesting it accounts for two-thirds of the 100 billion tons of methane produced globally by microorganisms each year [21,22,23,24,25,26]; consequently, acetate plays a crucial role in the global production of methane from organic matter. In coal seam environments, microbial community analyses and isotopic signatures have indicated the presence of acetoclastic methanogens alongside hydrogenotrophic and methylotrophic methanogens [27]. Only two identified genera of methanogens, Methanosarcina and Methanothrix, are capable of acetoclastic methanogenesis and thus are crucial to our current understanding of the global methane cycle [22]. While Methanosarcina barkeri, for example, is a generalist capable of producing methane from acetate as well as other substrates (e.g., CO2 + H2), Methanothrix soehngenii is an obligate acetoclastic methanogen that outcompetes Methanosarcina spp. at low acetate concentrations. Methanothrix-like spp. are thus thought to be the predominant acetoclastic methanogens in environmental settings where acetate is limited [28, 29]. While much is known independently about complex hydrocarbon degradation and methanogenesis, our present understanding of the microbial processes that link in situ metabolisms remains limited.
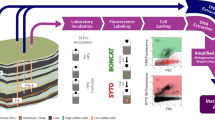
We determined potential metabolic linkages among microbial populations engaged in coal degradation, acetate production, and methanogenesis in the PRB Flowers-Goodale coal seam using a powerful combination of four primary techniques: (i) a nine-month in situ enrichment with crushed coal using a subsurface environmental sampler (SES) [30], (ii) bio-orthogonal non-canonical amino acid tagging (BONCAT) [31], (iii) fluorescently active cell sorting (FACS) [32], and (iv) genome-resolved metagenomics. Previous investigations have made major strides in surveying natural microbial communities by combining BONCAT-FACS with analyses of SSU rRNA gene sequences [33, 34], but genome-resolved metagenomics has yet to be performed on active cells following BONCAT-FACS and can provide biochemical predictions for active populations. In comparison to shotgun metagenomic sequencing, which sequences total community DNA and cannot discriminate between active, dead, and dormant organisms, sorting translationally active cells prior to metagenomic sequencing enables the identification of active microbial populations and associated genetic potential under relevant environmental conditions. We recovered high-quality, active metagenome-assembled genomes (MAGs) representing (i) a previously unidentified member of phylum Chlorobi with acetate-producing potential and (ii) a putative acetoclastic methanogen related to Methanothrix paradoxum. We hypothesize that these genomic populations (as well as members of the Geobacter and Bacteroidetes) interact in the degradation of aromatic coal byproducts and the subsequent production of methane from coal-derived acetate under fluctuating redox conditions.
Results & discussion
Recovery of high-quality, translationally active MAGs from an in situ coal enrichment
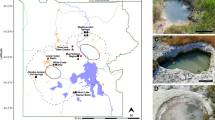
The SES was filled with crushed, subbituminous coal from the PRB and deposited at 115 m depth within a coal-bearing layer of the Flowers-Goodale coal seam at the U.S. Geological Survey (USGS) Birney Test Site (Fig. 1). Previous research demonstrated high concentrations (50 mg/L) of isotopically depleted methane (δ13C-CH4 = −67‰ versus VPDB) within this layer [6], indicating the presence of a microbial community engaged in CBM production. After a nine-month subsurface enrichment, the SES was retrieved maintaining in situ pressure and gaseous headspace conditions. We then performed BONCAT-FACS and sequenced the metagenome of translationally active and total sorted cell fractions. Metagenomic binning resulted in 24 metagenome assembled genomes (MAGs) from the translationally active fraction of the coal-enriched community (Supplementary Table 1) and 44 MAGs from the total cell fraction [35]. Two BONCAT-active genomic populations, Bin15 and Bin8, were recovered as high-quality MAGs with estimated completeness > 95% and estimated redundancy < 5% based on the detection of single copy genes for bacteria and archaea (Table 1). Robust phylogenomic analyses of concatenated archaeal ribosomal proteins indicated that Bin15 was a close neighbor to Methanothrix paradoxum NSM2 [36] within the Methanosarcinales order of methanogens (Fig. 2A), and henceforth will be referred to as “Methanothrix paradoxum PRB” for Powder River Basin. Consistent with this, M. paradoxum PRB had the highest genome-wide average nucleotide identity with M. paradoxum NSM2 at 77.6% when compared with other methanogens within and outside of the Methanosarcinales (Supplementary Table 2). M. paradoxum PRB had a genome size of 2.90 Mb, 50.7% G + C content and 2946 genes, similar to the type strain M. soehngenii GP6, which is 3.03 Mb with 51.9% G + C content and 2925 genes (Table 1).
We performed down-well incubation of sterile, crushed coal in the SES allowing for microbial colonization and retrieval under in situ pressure and anaerobic conditions. Samples were allocated into sterile gassed out serum bottles for addition of the bioorthogonal amino acid (HPG) in triplicate 24 hr incubations. We then sorted click-labeled BONCAT active cells (FAM Picolyl dye; Ex: 488 nm/Em: 530 nm) and total cells (SYTO59; Ex: 640 nm/Em: 655–685 nm) from each biological replicate. This was followed by DNA extraction, MDA amplification, sequencing, and analysis. The upper left coal seam stratigraphy panel was modified from Barnhart et al., 2016 [6].
Bacterial phylogenomic analysis indicated that high-quality, translationally active MAG Bin8 belonged to phylum Chlorobi and classified within the poorly understood OPB56 clade (Fig. 2B). Recently, Chlorobi groups were observed in situ via sequence analysis during long-term monitoring of an Australian coal seam post-stimulation for CBM production [37]. The Chlorobi phylum was first established to comprise the phototrophic Green Sulfur Bacteria, which is now considered class Chlorobea [38] and later revised to include the non-phototrophic class, Ignavibacteria [39]. OPB56 has been recognized as a third, class-level clade and was originally detected as a cluster of SSU rRNA gene sequence clones from Obsidian Pool in Yellowstone National Park [40]. Recent genomic discoveries by Hiras and colleagues [41] have confirmed the class-level OPB56 clade and, like the Ignavibacteria, OPB56 is non-phototrophic and contains genomes from thermophilic and non-thermophilic microbial populations. The “Chlorobi PRB” MAG (Bin8) was 3.34 Mb in length with 37.5% G + C content and a total of 2777 genes, in contrast to 2.67 Mb length, 56.0% G + C, and 2,363 genes for NICIL-2, the only other genome within the OPB56 clade that has been thoroughly evaluated [41].
The recovery of high-quality genomes for M. paradoxum PRB and Chlorobi PRB from the translationally active fraction suggests that these populations play key roles in the environment, though sequencing and/or amplification biases could also influence high genomic recoverability. We tested the hypothesis that these populations are ecologically relevant and detectable in coal seam environments by mapping quality-filtered short reads from four additional standard shotgun (non-BONCAT) metagenomes to the M. paradoxum PRB and Chlorobi PRB MAGs (Table 1). The four environmental metagenomes were from (i) the same well (FG11) but a different timepoint, (ii) a different well (FGP) from the same methane-producing coal seam, (iii) a non-methane-producing well (N11) also in the PRB [35], and (iv) a CBM-producing well (CX10) from the Surat Basin, Australia [42]. After normalizing for total sequence content, mean genomic coverage values for M. paradoxum PRB and Chlorobi PRB were 22X and 4X in FG11 and 153X and 0.25X in FGP, respectively, indicating that these populations exist naturally within the methane-producing Flowers-Goodale coal seam and may fluctuate in relative abundance based on environmental conditions. Moreover, a population corresponding to M. paradoxum PRB was also binned in a metagenome from a separate well in the Flowers-Goodale coal seam (FG09) and is presented in a companion study [35]. Genomic sequence of M. paradoxum PRB and Chlorobi PRB were also recovered from the total community sorted fraction (in addition to the translationally active fraction) after the in situ enrichment (i.e., Bin21 in Fig. 2A and Bin26 in Fig. 2B). In contrast, neither M. paradoxum PRB nor Chlorobi PRB recruited metagenomic reads from a non-methane-producing well (N11) in the PRB or a methane-producing well in the Surat Basin (CX10 [42], Table 1). This indicates that these populations may be endemic to high-CBM production wells in the PRB, but this hypothesis requires further testing as more metagenomic data are produced from coal seam environments. Due to the recovery of M. paradoxum PRB and Chlorobi PRB from multiple sequence sources (i.e., translationally active cell sorts, total cell sorts, binned environmental metagenomes, and mapped short reads) we hypothesize that these two populations play important and interconnected roles in the accumulation of methane in the PRB subsurface coal environment.
Metabolic properties of Chlorobi PRB
Metabolic comparisons between Chlorobi PRB and NICIL-2 [41] demonstrated consistent properties of the OPB56 class-level clade within phylum Chlorobi (Fig. 3; Supplementary Table 3). In contrast to the Green Sulfur Bacteria (class Chlorobea), OPB56 populations are not obligate anaerobes and do not possess genes involved in photosynthetic reactions (i.e., reaction centers [pscB, pscC, pscD], chlorosome envelope [csmABCDEFHIJX], bacteriochlorophyll a [fmoA]). In contrast, functional genes detected in Chlorobi PRB and NICIL-2 genomes suggest they are obligate heterotrophs with a facultative lifestyle capable of fermentation and aerobic respiration. NICIL-2 was previously enriched under oxic conditions and, like Chlorobi PRB, encodes multiple subunits for cytochrome c oxidases. Chlorobi PRB also encodes subunits of additional oxidases with varying binding affinities for oxygen, including cbb3-type cytochromes and bd-ubiquinols [43]. These observations suggest Chlorobi PRB may be adapted to respiring oxygen across a range of concentrations in subsurface coal seam environments. The cbb3-type oxidase, for example, is used by pathogenic proteobacteria to colonize anoxic zones in human tissue [44]. Chlorobi PRB also has the putative ability to perform anaerobic respiration via genes for membrane-bound nitrate reduction to ammonia (nrfAH [45]) and nitrous oxide reductase (nosZ [46]). In support of these findings, previous work using BONCAT-FACS on hot spring samples from Yellowstone National Park demonstrated that the OPB56 clade increased in SSU rRNA gene relative abundance when amended with oxygen or nitrate [34]. Putative respiration pathways are supported by the detection of complete electron transport pathways in both OPB56 genomes, and all genes were detected for an F-type proton-translocating ATP synthase. Like NICIL-2, Chlorobi PRB lacks genes for oxidation of sulfur compounds, distinguishing the OPB56 clade from members of class Chlorobea such as Chlorobaculum tepidum. Importantly, while Chlorobi PRB has high estimated genome completeness > 95%, the absence of genes could represent lack of genetic potential or genes that did not assemble with the MAG for methodological reasons. Therefore, missing functional properties inferred from gene absences are considered hypotheses that require further testing.
This figure is modified from a previous version published by Hiras and colleagues [41] for comparison with Chlorobi PRB. Colored enzyme numbers indicate which genome contained the respective gene (teal = Chlorobi PRB and NICIL-2; purple = Chlorobi PRB only; orange = NICIL-2 only; red = neither). Enzymes are numbered and are defined in Supplementary Table 3. [xyl xylose, xylu xyluose, ribu ribulose, rib ribose, glu glucose, pyru pyruvate; standard three-letter abbreviations are used for amino acids].
Further distinguishing OPB56 from the photosynthetic Green Sulfur Bacteria, both Chlorobi PRB and NICIL-2 encoded a complete TCA cycle and lacked genes for the rTCA cycle, which is the method by which members of the Chlorobea fix carbon. NICIL-2 has a complete glycolysis pathway while Chlorobi PRB is missing the enolase gene for the conversion of 2-phosphoglycerate to phosphoenolpyruvate. Both genomes have all additional genes necessary for gluconeogenesis. While NICIL-2 and Chlorobi PRB are both capable of fermentation, only Chlorobi PRB has the putative ability to produce acetate via a combination of the phosphotransacetylase (Pta) and acetate kinase (Ack) enzymes (discussed below). In contrast, NICIL-2 lacks the ack gene but has an alcohol dehydrogenase for the fermentative production of ethanol, which Chlorobi PRB does not. Carbon sources for NICIL-2 and Chlorobi PRB are primarily limited to simple, short-chain carbon compounds [41]; however, consistent with its recovery from a coal seam environment, the Chlorobi PRB genome also possessed genes for the anaerobic degradation of aromatic hydrocarbons such as phenylphosphate dehydrogenase (ppd), ethylbenzene dehydrogenase (ebd), and phenylethanol dehydrogenase (ped) (Table 2). By contrast, NICIL-2, which was not recovered from coal, only had the ped gene. Evidence for anaerobic hydrocarbon degradation in Chlorobi PRB together with the presence of anaerobic respiration genes (nrfAH, nosZ) may indicate that under anoxic conditions Chlorobi PRB respires hydrocarbons using oxidized nitrogen compounds [47, 48] and/or ferments hydrocarbons to acetate.
Both genomes from the OPB56 clade lack several biosynthesis pathways for amino acids, including leucine, valine, isoleucine, serine, phenylalanine, tryptophan, tyrosine, methionine, histidine, and proline. These results suggest that NICIL-2 and Chlorobi PRB likely rely on exogenous sources for many amino acids. Consistent with this, Reichart and colleagues demonstrated an increase SSU rRNA gene relative abundance of the OPB56 clade in enrichments amended with isoleucine [34], and the NICIL-2 and Chlorobi PRB genomes both have complete degradation pathways for isoleucine. Importantly, the missing biosynthesis pathway for methionine could enhance affinity for the synthetic amino acid HPG, which is a methionine analog. However, it should be noted that HPG levels used in short-term labeling incubations were low as previously described [33], and the Chlorobi sequences could be mapped back to unlabeled metagenomes from the environment. Finally, Chlorobi PRB and NICIL-2 encode all components of a complete flagellum complex, and Chlorobi PRB has two additional genes for flagellar chaperones, one that regulates flagellin polymerization (fliS [49]) and another that is essential to P ring formation (flgA [50]). These observations suggest that translationally active Chlorobi PRB may be motile in the subsurface coal seam, though genes for flagella are not always expressed, as is the case for cultures of Ignavibacterium album [39, 51]. Further investigations are needed to confirm suggested structures and functions based on gene detection/annotation.
Acetate production by Chlorobi PRB
As mentioned previously, Chlorobi PRB has the putative ability to produce acetate as a byproduct of fermentation using the Pta-Ack pathway. During fermentation, Pta catalyzes the replacement of CoA with a phosphate group and Ack subsequently cleaves the phosphate group, thereby releasing acetate while conserving energy in the form of 1 ATP [21]. Substrates for acetate production consist of many breakdown products of complex carbon sources, including glucose, propionate, and butyrate, as well as H2/CO2 in the case of homoacetogens. The importance of the Pta-Ack pathway in methanogenic environments plays a crucial role in coupling the breakdown of complex carbon to a substantial fraction of global methane production [22,23,24,25,26, 52]. In the Chlorobi PRB genome, ack and pta are adjacent genes on a contig with a length of ca. 10 kbp that contains seven genes. The neighboring gene to ack/pta is argE, which encodes acetylornithine deacetylase (COG0624) within the Zinc peptidase family (cl14876), an enzyme that catalyzes another acetate-producing reaction. Chlorobi PRB also has an additional copy of pta on a separate contig. These observations further support the hypothesis that the Chlorobi PRB population, which was translationally active after being enriched in situ on coal, may release acetate as a byproduct during the degradation of coal-derived aromatics.
Acetoclastic methanogenesis by Methanothrix paradoxum PRB
Only two genera of methanogens, Methanothrix and Methanosarcina, have been shown to use acetate as the sole source of carbon and energy during the production of methane [53]. Unlike Methanosarcina spp., which are generalists that can also grow on methylated compounds or hydrogen, Methanothrix spp. have no known substrates for methane production apart from acetate [22, 54]. Known Methanothrix spp. have extremely high affinity for acetate and can outcompete Methanosarcina spp. by growing at lower acetate concentrations in many environments. While Methanosarcina spp. use the previously discussed Pta-Ack pathway in reverse to activate acetate for methane production, Methanothrix spp. instead use acetyl-CoA synthetase (acs). M. paradoxum PRB from the present study, M. paradoxum NSM2 [36], and M. soehngenii GP6 [54] all harbor genes consistent with the latter type of acetoclastic methanogenesis (Table 2; Supplementary Table 4), including acs for acetate activation, carbon monoxide dehydrogenases (cdh) for cleavage of carbon groups and oxidation of CO, methyltransferases (mtr) for activation of the methyl group, and methyl coenzyme M reductase (mcr) for the final reduction step that produces methane [20, 22]. Four adjacent copies of acs existed on a 24,695-kbp contig and a fifth copy was detected on a separate contig. The cdh operon was ordered alpha, epsilon, beta, CooC1 Ni-accessory protein, delta, gamma, and was present on a single contig spanning 41,179 kbp. The mcr operon was on a large contig >40 kbp in length and consisted of subunits ordered beta, D, gamma, alpha. In addition to methanogenesis from acetate, hidden Markov model (HMM) scans for anaerobic hydrocarbon degradation proteins [55] detected putative protein sequences for breaking down phenylethanol (ped), toluene and xylene (bss), and phenol (pps). These observations suggest that, in addition to producing methane directly from acetate, M. paradoxum PRB may directly or indirectly use certain coal byproducts as additional sources of carbon and/or energy. These results coincide with the recent discovery of a different methanogen, Methermicoccus shengliensis, which has the ability to utilize methyl-groups from methoxylated aromatic compounds [56].
Considerations of oxygen tolerance and usage
Several important findings challenge the classical understanding that all methanogens are strict anaerobes. Possible aerotolerant methanogens have been identified in methanogenic ecosystems such as rice paddy fields, arid soils, and anaerobic digesters (e.g., [57,58,59]). Several more observations of possible oxygen tolerance come specifically from the acetoclastic Methanothrix genus. First, the original cultivations of M. soehngenii GP6 demonstrated that growth could be attained starting from aerobic samples and on sewage exposed to pure oxygen for up to 48 h [54]. Second, Jetten and colleagues [22, 28] purified and characterized the Cdh enzyme from M. soehngenii, which was determined to be “completely insensitive to molecular oxygen,” in contrast to the same enzyme from Methanosarcina barkeri which irreversibly decreased in activity by 90% after trace oxygen exposure [60]. Phylogenetic analysis of the Cdh enzyme from M. paradoxum PRB confirms placement as a neighbor to the Cdh from M. soehngenii, together forming a separate cluster from the Methanosarcinales Cdh group (Supplementary Fig. 1). Finally, Angle et al. [36] observed that methane production increased by up to an order of magnitude in oxygenated wetland soils compared to anoxic soils and methanogenesis was attributed primarily to acetoclastic M. paradoxum. In the present study, SES oxygen measurements in the FGP well ranged from 0.25 to 1.09 mole % (n = 5) (Supplementary Table 5). Although we cannot rule out potential oxygen contamination from sampling or analysis, previous metagenomic analyses of coal seam environments have indicated the importance of aerobic or microaerophilic metabolisms in such environments [61]. The observed oxygen fluctuations are consistent with the wide-ranging potential for energy conservation observed in Chlorobi PRB, which encodes oxygenases with varying binding affinities (high to low oxygen concentrations), nitrate reductases, and fermentation enzymes.
Remarkably, the M. paradoxum PRB genome harbored an extradiol dioxygenase (elh) gene from the LigB superfamily for the aerobic degradation of phenylacetate [62]. The presumptive protein sequence was recovered with a 40.8% amino acid similarity in the HMM scan, with an expect (e) value of 3.0 × 10−25 (e value of 5.6 × 10−86 for match to COG2078, Supplementary Table 4A). We ruled out sequence contamination by comparing the full 36-kb contig containing the elh gene to the NCBI non-redundant sequence database and observed a closest nucleotide sequence match (77.76% identity, e value = 0) to M. soehngenii GP6. Consistent with this, M. soehngenii GP6 and M. paradoxum NSM2 both had copies of elh as well. Further support for phenylacetate metabolism in M. paradoxum PRB was observed in a neighboring gene for phenylacetate-coenzyme A ligase (COG1451, adenylate-forming domain family), which occurred just three genes downstream of elh for aerobic phenylacetate degradation (Supplementary Table 6). Phenylacetate has been demonstrated as a key intermediate in the conversion of organic matter to methane by accumulation in peat soil enrichments when methanogenesis was inhibited; in some inhibition experiments, phenylacetate accumulated to even higher concentrations than acetate [25]. Finally, growth and methane production were observed for M. soehngenii in the presence of acetate and phenylacetate, although not on phenylacetate alone [54]. Due to the (i) apparent relationship between phenylacetate and acetoclastic methanogenesis, (ii) the unique oxygen-tolerating characteristics of Methanothrix spp., (iii) the presence of the elh dioxygenase for phenylacetate degradation in M. paradoxum PRB, and (iv) the observed fluctuating redox conditions of the Flowers-Goodale coal seam, we hypothesize that M. paradoxum PRB may use trace oxygen for ring cleavage of coal-derived phenylacetate during or as an alternative (and/or supplement) to the production of methane from acetate. Further research such as methanogenic cultivations under oxic/suboxic conditions and purifications of the novel Elh enzyme would be needed to test this hypothesis.
Biological process for CBM production
The recovery of two high-quality MAGs with ostensibly related putative metabolisms (i.e., acetoclastic methanogenesis in M. paradoxum PRB and acetate production in Chlorobi PRB) indicated the importance of acetate as an intermediate substrate during CBM production. We scanned the lower quality, translationally active MAGs for the putative ability to produce acetate via the Ack/Pta pathway and identified members of the Bacteroidetes and Geobacter with ack/pta genes (Table 1; Supplementary Table 1). However, the Ack/Pta pathway can be used in reverse during acetate activation to acetyl-CoA. Acetate consumption via Ack/Pta has been demonstrated for Geobacter sulfurreducens [63], suggesting that Geobacter in the PRB may compete with M. paradoxum PRB for acetate dependent upon availability of potential electron acceptors. Further supporting this hypothesis, Beckmann et al. used DNA stable isotope probing to demonstrate carbon assimilation from acetate by Geobacter spp. and methanogens together in the same methane-producing coal seam in Australia [37]. In addition to reverse Ack/Pta, pyruvate:ferredoxin oxidoreductase (PFOR) has recently been suggested by in silico analysis to generate pyruvate from acetate in a single step in G. sulfurreducens [64], representing another pathway for acetate utilization. PFOR is common among anaerobic microorganisms for the reversible oxidation of pyruvate to acetyl-CoA [65, 66]. All three bacterial MAGs (Geobacter PRB, Chlorobi PRB, and Bacteroidetes PRB) encode at least one PFOR as well as Ack/Pta; however, given the demonstrated reversibility of the respective reactions, directionality is difficult to predict for in situ conditions. Recent work has shown that the direction of the Ack/Pta pathway in Escherichia coli is determined by thermodynamic controls [67], suggesting redox conditions and/or metabolite availability in the Flowers-Goodale coal seam may ultimately determine whether these bacterial populations consume or produce acetate for methanogenesis by M. paradoxum PRB.
Deduced polypeptide sequences for Bacteroidetes and Geobacter MAGs were scanned for aerobic and anaerobic hydrocarbon degradation enzymes. Similar to M. paradoxum PRB, both Bacteroidetes PRB and Geobacter PRB encoded the Elh enzyme suggesting aerobic phenylacetate degradation, and all elh genes were on contigs taxonomically confirmed by total nucleotide matches to the same taxonomic groups. In terms of anaerobic hydrocarbon metabolism, Bacteroidetes PRB and Geobacter PRB both had multiple copies of the gene encoding Ped for the degradation of phenylethanol. Bacteriodetes PRB, like Chlorobi PRB, encoded a phenylphosphate carboxylase (Ppc), while Geobacter PRB encoded the phenylphosphate synthase (Pps, like M. paradoxum PRB) and the Ebd for the degradation of ethylbenzene (like Chlorobi PRB). Members of the Bacteroidetes have been associated with complex carbon turnover in suboxic to anoxic environments sometimes associated with methanogenesis [68, 69]. Geobacter sequences and/or organisms have been observed in different environments associated with the turnover of recalcitrant carbon and/or methanogenesis. For example, in recent studies, Geobacter were shown to be increased with biochar samples and increased methanogenesis [70], correlated to decreased polyphenolics/polycyclic aromatics in methanogenic rice paddy soils [71], and shown to catalyze the turnover of organic matter associated with Fe (hydr)oxides [72].
Our genome-resolved analyses of the translationally active community in the PRB subsurface reveal a conceptual model describing important populations and their associated biochemical capacities that contribute to microbial CBM production (Fig. 4). By incubating coal down-well in an SES for nine months and allowing establishment of a coal-dependent microbial community under in situ methanogenic conditions, the coal-community was secured at depth before retrieval, and then retrieved to the surface in a sealed chamber. M. paradoxum PRB is likely a key methanogen in the PRB subsurface with the genomic potential to convert acetate to methane, and this population apparently becomes active in the presence of crushed coal in situ. Sources of acetate are likely derived from Chlorobi PRB via the Pta-Ack pathway, with possible additional contributions coming from Bacteroidetes and Geobacter populations, though these populations may also consume acetate depending on environmental conditions. Together, all four translationally active populations (M. paradoxum PRB, Chlorobi PRB, Bacteroidetes PRB, and Geobacter PRB) have the combined genomic potential for the anaerobic degradation of ethylbenzene, phenylphosphate, phenylethanol, toluene, xylene, and phenol. M. paradoxum PRB, Bacteroidetes PRB, and Geobacter PRB have the additional potential to break down phenylacetate under micro-aerobic conditions. Finally, certain hydrocarbon degradation enzymes are linked by related pathways, such as Ebd and Ped, which catalyze conversions of ethylbenzene to phenylethanol and subsequently phenylethanol to acetophenone, respectively. All four MAGs possess the ped gene but only Chlorobi PRB and Geobacter PRB possess strong matches to the ebd gene. These data provide insights into a coalbed methane community that is likely metabolically interconnected in which hydrocarbon conversions by certain community members stimulate downstream conversions by others.
Translationally active MAGs in the PRB harbor the putative ability to degrade a variety of aromatics, including ethylbenzene, phenylethanol, phenol, phenylphosphate, toluene, xylene, and phenylacetate. Arrows representing genes or deduced enzymes are colored by the host microbial population (orange = Chlorobi PRB, purple = Geobacter PRB, blue = Bacteroidetes PRB, teal = M. paradoxum PRB) and indicate that either carbon or energy may be derived from the putative reaction. Dashed lines indicate putative oxygen-consuming reactions. Detailed metabolic potential of the Chlorobi PRB MAG is presented in Fig. 3. [ebd = ethylbenzene dehydrogenase, ped = phenylethanol dehydrogenase, pps = phenylphosphate synthase, ppc = phenylphosphate carboxylase, bss = putative benzylsuccinate synthase, ack = acetate kinase, pta = phosphotransacetylase, acs = acetyl CoA synthase, cdh = carbon monoxide dehydrogenase, mtr = methyltransferase, mcr = methyl CoM reductase, elh = extradiol dioxygenase: LigB superfamily: homoprotocatechuate].
Conclusions
Subsurface environments associated with different forms of hydrocarbons account for up to 1013 metric tons of carbon globally that can be ultimately recycled back to CO2 and CH4 [73]. Investigations into how microbial communities interact to complete different stages of carbon remineralization in these environments—from initial interactions and degradation of complex aromatics to ultimately the production of methane and carbon dioxide from precursor metabolites—can provide insight for potential contributions to the global carbon cycle with impacts ranging from climate change to the energy sector. Unfortunately, very little is known regarding many of the steps associated with the degradation of recalcitrant hydrocarbons in the subsurface under in situ conditions as these environments are extremely difficult to sample and many of the associated microorganisms are not known and/or not in cultivation. To this end, we used a unique SES device to allow an ecologically relevant community to establish under in situ conditions (enriched on crushed coal in the subsurface), and then used novel techniques in activity-based metagenomics to identify translationally active members of the microbial community. Our observations indicate that in this coal-bearing subsurface ecosystem, specific microbial populations facilitate the biological conversion of coal degradation products to methane using acetate as a key intermediate. Genomic analyses of Chlorobi PRB (and perhaps Bacteroidetes PRB and Geobacter PRB) suggested putative abilities to degrade aromatic hydrocarbons (anaerobically or aerobically) and produce acetate for the subsequent production of methane by the putative acetoclastic methanogen, Methanothrix paradoxum PRB. Consequently, these microbial populations may play crucial roles cycling carbon in a shallow subsurface coal seam environment that contributes to the conversion of coal to methane gas.
Materials and methods
Down-well sampling, BONCAT incubations
The following methods are also presented in a companion study that is a broad-scope analysis of microbial coal degradation processes (e.g., fumarate addition, biosurfactant production) at multiple sites in the PRB under varying sulfate conditions [35]. In September 2017, an SES (Patent # US10704993B2) was loaded with UV-sterile crushed coal, lowered by cable to a depth of 115 m in the FG11 well in the PRB (at the U.S. Geological Survey’s Birney site) [6], and opened by a control box at the surface. After nine months of down-well incubation the SES was closed, forming a gas-tight chamber, and retrieved to the surface. 10-ml SES slurries were extracted through a Swagelok device (Solon, Ohio, USA) and anoxically transferred into sterile balch tubes (95% N2, 5% CO2) in triplicate. L-homopropargylglycine (HPG, Click Chemistry Tools, Scottsdale, Arizona, USA) was prepared in sterile degassed water (DEPC diethyl pyrocarbonate treated filter sterilized water, pH 7) and added to each replicate at a final concentration of 250 μM. Higher HPG concentrations were used compared to previous studies (e.g., [33, 34]) to overcome loss of the bioorthogonal amino acid due to sorption to porous coal. To control for sorting artifacts samples were prepared the same way, except did not have HPG added (HPG negative control). All samples were incubated in the dark at 20 °C for 24 h (compared to an in situ temperature range of 16–18 °C, Supplementary Table 5). We note that the BONCAT methodology requires relatively short incubation times (<48 h [33, 34]) to prevent over-labeling and/or cross-labeling. In this case a 24-hr incubation was selected based on experimental verification of identifiable cells by fluorescence microscopy and in attempt to minimize bottle effects for the subsamples removed from the SES. Following incubation, cells were removed from coal according to the protocol described by Couradeau et al. [33]. Briefly, 1 ml of slurry was removed and added to Tween® 20 at a final concentration of 0.02% (Sigma–Aldrich) in phosphate saline buffer (1X PBS). Samples were then vortexed at maximum speed for 5 min followed by centrifugation at 500 xG for 5 min [33]. The supernatant was removed, filtered through a 40-µm strainer, and spun at 14,000 xG to concentrate detached cells. The cell pellet was immediately cryopreserved at −20 °C in a sterile 55% glycerol TE (11X) solution.
Fluorescent labeling, cell sorting, amplification, and metagenomic sequencing
The click reaction buffer consisted of copper sulfate (CuSO4 100 µM final concentration), tris-hydroxypropyltriazolylmethylamine (THPTA, 500 µM final concentration), and FAM picolyl azide dye (5 µM final concentration) [32]. For the click reaction, each sample (200 µl) was placed on a 25-mm 0.2-µm polycarbonate filter resting on a microscope slide, and 80 µl of BONCAT click reaction was added before covering with a coverslip. The BONCAT click reaction consisted of 5 mM Sodium Ascorbate, 5 mM Aminoguanidine HCl, 500 µM THPTA, 100 µM CuSO4, and 5 µM FAM picolyl azide in 1X phosphate buffered saline. Incubation time was 30 min, followed by three washes in 20 ml of 1X PBS for 5 min each. Cells were recovered from the filter by vortexing in 0.02% Tween for 5 min, and then stained using 0.5 µM SYTOTM59 (ThermoFisher Scientific, Invitrogen, Eugene Oregon, USA) DNA stain.
For cell sorting a BD-InfluxTM (BD Biosciences, San Jose, California, USA) specifically configured to capture total cells (SYTOTM59 [excitation = 622 nm, emission = 645 nm]) in the red region of a 640-nm laser and BONCAT active cells (FAM picolyl azide dye [excitation = 490 nm/emission = 510 nm]) in the green region of a 488-nm blue laser. The total cell population was gated for BONCAT positivity by comparing the 530/40 BP fluorescence off a 488-nm laser against an HPG negative control that had undergone the same click reaction. Two fractions (total cells and BONCAT active cells) were sorted from each replicate sample. The first faction consisted of the DNA + cells, and the second only contained BONCAT + cells as determined by comparison to the HPG negative control. Fractions were sorted into 394 well plates, and for each fraction 5,000 cells were collected into 4 wells and 300 cells were collected into 20 well. Following sorting plates were frozen at −80 °C until further processing.
Cells were pelleted from wells containing 5000 cells via centrifugation (6000 xG for 1 hr at 10 °C), followed by removal of the supernatant and a brief inverted spin at 6 xG. This step was necessary to avoid interference with subsequent whole genome amplification reaction chemistry. Wells containing only 300 sorted cells were not pelleted, rather were directly lysed and amplified using 5 µl WGAX reactions following optimized conditions [74]. Briefly, cells were lysed in 650 nl lysis buffer for 10 min at room temperature. The lysis buffer consisted of 300 nl TE + 350 nl of 400 mM KOH, 10 mM EDTA, and 100 mM DTT. Lysis reactions were neutralized by the addition of 350 nl of 315 mM HCl in Tris-HCl. Whole genome amplification reactions were brought to 5 µl with final concentrations of 1X EquiPhi29 reaction buffer (ThermoFisher), 0.2 U/µl EquiPhi29 polymerase (Thermo), 0.4 mM dNTPs, 50 µM random heptamers, 10 mM DTT, and 0.5 µM SYTO13. Plates were incubated at 45 °C for 13 hr. Libraries for metagenomic sequencing were created using the Nextera XT v2 kit (Illumina) with 12 rounds of PCR amplification. All volumes and inputs to Nextera reactions were reduced 10-fold from the manufacturer’s recommendations. Libraries were sequenced 2 × 150 bp mode on the Nextseq platform (Illumina).
Metagenomic assembly and binning
Raw metagenomic short reads were quality filtered using illumina-utils [75] (v1.0) with default parameters. Technical sequencing replicates were coassembled with MEGAHIT [76] (v1.2.9) for each of three biological replicates for both BONCAT-active and TOTAL sorted cells, resulting in six metagenome assemblies. These three replicate assemblies from BONCAT-active and the additional three replicates from TOTAL cell fractions were further coassembled via MEGAHIT, resulting in a single metagenome assembly for BONCAT and another for TOTAL. Assembled sequences were filtered at a minimum length cutoff of 5000 bp and binned in anvi’o [77] (v6) based on tetranucleotide frequencies with a scaffold split size of 20,000 bp. Genome bins were scanned for single copy genes to estimate completeness and redundancy and bins were refined until estimated redundancy was < 10%. PyANIp [78] (v0.2.10) was used to compare genomic bins between BONCAT and TOTAL assemblies, and bins with alignment lengths > 85% and average nucleotide identities (ANI) greater than 95% were considered the same microbial population recovered from both BONCAT and TOTAL. Anvi’o was used to summarize additional genomic information of all bins, including % G + C, N50, number of contigs, and cumulative sequence length.
Environmental detection of active genome bins
Three months after the BONCAT-metagenomics experiment, additional coal-enriched samples were retrieved from the FG11 well and FGP—a nearby, hydrologically connected well—for standard shotgun metagenomic sequencing. As a background control, samples were also collected from a non-methane-producing coal seam well (N11). DNA was isolated from these samples using the MP Biomedical ProDNA Spin Kit for Soil according to the manufacturer’s protocol. Metagenomic sequencing was performed by the U.S Department of Energy Joint Genome Institute and raw reads were quality-filtered as above. Bowtie2 [79] (v2.2.6) was used to map FG11, FGP, and N11 short reads against BONCAT-active genome bins to retrieve sequence coverage values for these bins directly from the environment. We also downloaded metagenomic data from a methane-producing coal seam in the Surate Basin, Australia (CX10), for additional comparison (NCBI SRA: SRX1122679) [42]. Total sequence content in quality-filtered fastq files from FG11 (19.5 Gb), FGP (25.2 Gb), N11 (11.5 Gb), and CX10 (8.9 Gb) was used to normalize coverage values for comparisons across sites.
Phylogenetic analyses and taxonomic designations
For genomic bins with the highest estimated completeness (i.e., M. paradoxum PRB, Chlorobi PRB) as well as taxonomically related reference genomes, 16 ribosomal protein sequences were extracted via anvi’o and concatenated in the following order: L27A, S10, L2, L3, L4, L18p, L6, S8, L5, L24, L14, S17, S3_C, L22, S19, L16RP. Muscle [80] (v3.8.31) was used to align concatenated ribosomal protein sequences with eight maximum iterations. Phylogenetic analyses of aligned concatenated proteins were performed for archaea and bacteria with MrBayes [81] (v3.2.6) using a fixed aa model, empirical aa frequencies, eight gamma distribution categories, eight parallel chains, and a burn-in fraction of 0.25. We ran 100,000 generations for the archaeal analysis and 1000,000 generations for the bacteria, resulting in standard deviations in split frequencies of 0.000235 and 0.000000, respectively. Taxonomies of the other bins were determined by nucleotide sequence comparisons with BLASTn [82] to the NCBI non-redundant database [83]. Bacteroidetes_PRB_Bin13 and Geobacter_PRB_Bin11 bins did not contain enough ribosomal proteins for phylogenetic analysis and were instead assigned rank level taxonomy based on nearest matches to each contig within each bin. The majority of contigs in Bin 11 (82 of 151) had strong matches to members of the Geobacter, while the remaining hits were closely related members of the Deltaproteobacteria phylum. In contrast, Bin 13 could not be resolved beyond the phylum level. Only 79 of the 126 contigs in Bin 13 had hits to the NCBI database and those hits were scattered amongst diverse members within the Bacteroidetes phylum (e.g., Flavobacterium, Draconibacterium, Sphingobacteriaceae).
An additional Bayesian phylogenetic tree was calculated for the Cdh enzyme subunit alpha (approximately 800 aa in length) from methanogens. First, Cdh sequences were aligned with MAFFT [84] (maxiterate 1000, localpair) and trimmed with BMGE [85] (BLOSUM30). We used MrBayes [81] to calculate the Bayesian tree with a mixed amino acid model. The standard deviation of split frequencies was 0.0000 after 1000,000 iterations and all posterior probabilities were 1.00.
Metabolic analysis
Prodigal [86] was used in the anvi’o platform to identify open reading frames within genomic sequence. Initial functional annotations were conducted using the KEGG database [87] with deduced amino acid. METABOLIC [88] was also used for identification of functional genes related to major biogeochemical cycles. The Chlorobi PRB genome was further examined in direct comparison to the NICIL-2 genome using EC pathway annotations focused on important attributes of the Chlorobi phylum outlined by Hiras et al. [41]. We used HMM scans of deduced protein sequences against the AromaDeg database [89] to uncover enzymes associated with aerobic aromatic hydrocarbon degradation. Similarly, for anaerobic conversions of aromatic hydrocarbons we scanned deduced proteins for enzymes in the AnHyDeg database [55]. For genes of interest (e.g., related to methane metabolism, Pta-Ack pathway, phenylacetate degradation) we examined host contigs to confirm taxonomic calls by using BLASTn [82] against the NCBI non-redundant database [83]. Contigs were further examined for neighboring genes related to the same microbial process (e.g., subunits of the same enzyme).
Geochemical analyses
Water samples analyzed for pH, temperature, CH4, and δ13C-CH4 were collected with a Grundfos submersible pump after three wellbore volumes were pumped and field properties stabilized and were analyzed as previously described [6, 27]. Samples for O2 and other gases were collected with an SES. The internal substrate chamber of the SES was removed, and the SES was slowly dropped down-well to the center of the well screen depth to collect gas and water samples. The SES remained open at the center of the well screen for several minutes before closing the SES and retrieving to the surface. Gas concentrations were measured using a headspace equilibration technique developed by Isotech Laboratories, Inc. (a Stratum Reservoir brand) as described previously [90] with detailed analysis information available through Isotech Laboratories (www.isotechlabs.com). Acetate concentrations were measured by previously reported methods [91].
Data availability
Genomic sequence data associated with Total-sorted and BONCAT-sorted cells are available on the Integrated Microbial Genomes & Microbiomes (IMG) site under GOLD Study ID Gs014100. High quality MAGs for Methanothrix paradoxum PRB and Chlorobi PRB were submitted to JGI under GOLD Analysis IDs Ga0496496 and Ga0496497, respectively. Environmental metagenomes for FG11 and FGP wells are available on IMG under GOLD Project IDs Gp0406117 and Gp0406116, respectively.
References
Colosimo F, Thomas R, Lloyd JR, Taylor KG, Boothman C, Smith AD, et al. Biogenic methane in shale gas and coal bed methane: a review of current knowledge and gaps. Int J Coal Geol. 2016;165:106–20.
Strąpoć D, Mastalerz M, Dawson K, Macalady J, Callaghan AV, Wawrik B, et al. Biogeochemistry of microbial coal-bed methane. Annu Rev Earth Planet Sci. 2011;39:617–56.
Barnhart EP, Davis KJ, Varonka M, Orem W, Cunningham AB, Ramsay BD, et al. Enhanced coal-dependent methanogenesis coupled with algal biofuels: Potential water recycle and carbon capture. Int J Coal Geol. 2017;171:69–75.
Huang Z, Sednek C, Urynowicz MA, Guo H, Wang Q, Fallgren P, et al. Low carbon renewable natural gas production from coalbeds and implications for carbon capture and storage. Nat Commun. 2017;8:568.
Pachauri RK, Allen MR, Barros VR, Broome J, Cramer W, Christ R, et al. Climate Change 2014: Synthesis Report. Contribution of Working Groups I, II and III to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. 2014. IPCC, Geneva, Switzerland.
Barnhart EP, Weeks EP, Jones EJP, Ritter DJ, McIntosh JC, Clark AC, et al. Hydrogeochemistry and coal-associated bacterial populations from a methanogenic coal bed. Int J Coal Geol. 2016;162:14–26.
Ritter D, Vinson D, Barnhart E, Akob DM, Fields MW, Cunningham AB, et al. Enhanced microbial coalbed methane generation: a review of research, commercial activity, and remaining challenges. Int J Coal Geol. 2015;146:28–41.
Zhuravlev YN, Porokhnov AN. Computer simulation of coal organic mass structure and its sorption properties. Int J Coal Sci Technol. 2019;6:438–44.
Sondreal EA, Wiltsee GA. Low-rank coal: its present and future role in the United States. Annu Rev Energy Environ. 1984;9:473–99.
Zhang R, Liu S, Bahadur J, Elsworth D, Wang Y, Hu G, et al. Changes in pore structure of coal caused by coal-to-gas bioconversion. Sci Rep. 2017;7:3840.
Lu Y, Chai C, Zhou Z, Ge Z, Yang M. Influence of bioconversion on pore structure of bituminous coal. Asia-Pac J Chem Eng. 2020;15:e2399.
Glombitza C, Mangelsdorf K, Horsfield B. A novel procedure to detect low molecular weight compounds released by alkaline ester cleavage from low maturity coals to assess its feedstock potential for deep microbial life. Org Geochem. 2009;40:175–83.
Jones EJP, Voytek MA, Corum MD, Orem WH. Stimulation of methane generation from nonproductive coal by addition of nutrients or a microbial consortium. Appl Environ Microbiol. 2010;76:7013–22.
Vinson DS, Blair NE, Ritter DJ, Martini AM, McIntosh JC. Carbon mass balance, isotopic tracers of biogenic methane, and the role of acetate in coal beds: Powder River Basin (USA). Chem Geol. 2019;530:119329.
Bapteste E, Brochier C, Boucher Y. Higher-level classification of the Archaea: evolution of methanogenesis and methanogens. Archaea. 2005;1:353–63.
Paul K, Nonoh JO, Mikulski L, Brune A. “Methanoplasmatales,” Thermoplasmatales-related archaea in termite guts and other environments, are the seventh order of Methanogens. Appl Environ Microbiol. 2012;78:8245–53.
Vanwonterghem I, Evans PN, Parks DH, Jensen PD, Woodcroft BJ, Hugenholtz P, et al. Methylotrophic methanogenesis discovered in the archaeal phylum Verstraetearchaeota. Nat Microbiol. 2016;1:16170.
Borrel G, Adam PS, McKay LJ, Chen L-X, Sierra-García IN, Sieber CMK, et al. Wide diversity of methane and short-chain alkane metabolisms in uncultured archaea. Nat Microbiol. 2019;4:603–13.
McKay LJ, Dlakić M, Fields MW, Delmont TO, Eren AM, Jay ZJ, et al. Co-occurring genomic capacity for anaerobic methane and dissimilatory sulfur metabolisms discovered in the Korarchaeota. Nat Microbiol. 2019;4:614–22.
Liu Y, Whitman WB. Metabolic, phylogenetic, and ecological diversity of the methanogenic archaea. Ann N. Y Acad Sci. 2008;1125:171–89.
Ferry JG. Acetate kinase and phosphotransacetylase. Methods Enzymol. 2011;494:219–31.
Jetten MSM, Stams AJM, Zehnder AJB. Methanogenesis from acetate: a comparison of the acetate metabolism in Methanothrix soehngenii and Methanosarcina spp. FEMS Microbiol Rev. 1992;8:181–97.
Zinder SH. Physiological Ecology of Methanogens. In: Ferry JG (ed). Methanogenesis: Ecology, Physiology, Biochemistry & Genetics. 1993. Springer US, Boston, MA, pp. 128–206.
Ferry JG. How to make a living by exhaling methane. Annu Rev Microbiol. 2010;64:453–73.
Kotsyurbenko OR, Chin K-J, Glagolev MV, Stubner S, Simankova MV, Nozhevnikova AN, et al. Acetoclastic and hydrogenotrophic methane production and methanogenic populations in an acidic West-Siberian peat bog. Environ Microbiol. 2004;6:1159–73.
Conrad R. Contribution of hydrogen to methane production and control of hydrogen concentrations in methanogenic soils and sediments. FEMS Microbiol Ecol. 1999;28:193–202.
Schweitzer H, Ritter D, McIntosh J, Barnhart E, Cunningham AB, Vinson D, et al. Changes in microbial communities and associated water and gas geochemistry across a sulfate gradient in coal beds: Powder River Basin, USA. Geochim Cosmochim Acta. 2019;245:495–513.
Jetten MS, Stams AJ, Zehnder AJ. Isolation and characterization of acetyl-coenzyme A synthetase from Methanothrix soehngenii. J Bacteriol. 1989;171:5430–5.
Gujer W, Zehnder AJB. Conversion Processes in Anaerobic Digestion. Water Sci Technol; Lond. 1983;15:127–67.
Barnhart EP, Ruppert L, Hiebert R, Smith H, Schweitzer H, Clark A, et al. Injection of Deuterium and Yeast Extract at USGS Birney Field Site, Powder River Basin, Montana, USA, 2016–2020. US Geological Survey Data Release 2021.
Hatzenpichler R, Scheller S, Tavormina PL, Babin BM, Tirrell DA, Orphan VJ. In situ visualization of newly synthesized proteins in environmental microbes using amino acid tagging and click chemistry. Environ Microbiol. 2014;16:2568–90.
Hatzenpichler R, Connon SA, Goudeau D, Malmstrom RR, Woyke T, Orphan VJ. Visualizing in situ translational activity for identifying and sorting slow-growing archaeal- bacterial consortia. Proc Natl Acad Sci. 2016;113:E4069–78.
Couradeau E, Sasse J, Goudeau D, Nath N, Hazen TC, Bowen BP, et al. Probing the active fraction of soil microbiomes using BONCAT-FACS. Nat Commun. 2019;10:2770.
Reichart NJ, Jay ZJ, Krukenberg V, Parker AE, Spietz RL, Hatzenpichler R. Activity-based cell sorting reveals responses of uncultured archaea and bacteria to substrate amendment. ISME J. 2020;14:2851–61.
Schweitzer H, Smith H, Barnhart EP, McKay L, Gerlach R, Cunningham AB, et al. Subsurface Hydrocarbon Degradation Strategies in Low- and High-Sulfate Coal Seam Communities Identified with Activity-Based Metagenomics. bioRxiv 2021. https://doi.org/10.1101/2021.08.26.457739.
Angle JC, Morin TH, Solden LM, Narrowe AB, Smith GJ, Borton MA, et al. Methanogenesis in oxygenated soils is a substantial fraction of wetland methane emissions. Nat Commun. 2017;8:1567.
Beckmann S, Luk AWS, Gutierrez-Zamora M-L, Chong NHH, Thomas T, Lee M, et al. Long-term succession in a coal seam microbiome during in situ biostimulation of coalbed-methane generation. ISME J. 2019;13:632–50.
Imhoff JF. Phylogenetic taxonomy of the family Chlorobiaceae on the basis of 16S rRNA and fmo (Fenna-Matthews-Olson protein) gene sequences. Int J Syst Evol Microbiol. 2003;53:941–51.
Iino T, Mori K, Uchino Y, Nakagawa T, Harayama S, Suzuki K-I. Ignavibacterium album gen. nov., sp. nov., a moderately thermophilic anaerobic bacterium isolated from microbial mats at a terrestrial hot spring and proposal of Ignavibacteria classis nov., for a novel lineage at the periphery of green sulfur bacteria. Int J Syst Evol Microbiol. 2010;60:1376–82.
Hugenholtz P, Pitulle C, Hershberger KL, Pace NR. Novel division level bacterial diversity in a Yellowstone hot spring. J Bacteriol. 1998;180:366–76.
Hiras J, Wu Y-W, Eichorst SA, Simmons BA, Singer SW. Refining the phylum Chlorobi by resolving the phylogeny and metabolic potential of the representative of a deeply branching, uncultivated lineage. ISME J. 2016;10:833–45.
Evans PN, Parks DH, Chadwick GL, Robbins SJ, Orphan VJ, Golding SD, et al. Methane metabolism in the archaeal phylum Bathyarchaeota revealed by genome-centric metagenomics. Science. 2015;350:434–8.
Morris RL, Schmidt TM. Shallow breathing: bacterial life at low O(2). Nat Rev Microbiol. 2013;11:205–12.
Pitcher RS, Watmough NJ. The bacterial cytochrome cbb3 oxidases. Biochim Biophys Acta. 2004;1655:388–99.
Simon J, Pisa R, Stein T, Eichler R, Klimmek O, Gross R. The tetraheme cytochrome c NrfH is required to anchor the cytochrome c nitrite reductase (NrfA) in the membrane of Wolinella succinogenes. Eur J Biochem. 2001;268:5776–82.
Orellana LH, Rodriguez-R LM, Higgins S, Chee-Sanford JC, Sanford RA, Ritalahti KM, et al. Detecting nitrous oxide reductase (NosZ) genes in soil metagenomes: method development and implications for the nitrogen cycle. mBio. 2014;5:e01193–14.
Coates JD, Chakraborty R, Lack JG, O’Connor SM, Cole KA, Bender KS, et al. Anaerobic benzene oxidation coupled to nitrate reduction in pure culture by two strains of Dechloromonas. Nature. 2001;411:1039–43.
Chakraborty R, Coates JD. Anaerobic degradation of monoaromatic hydrocarbons. Appl Microbiol Biotechnol. 2004;64:437–46.
Muskotál A, Király R, Sebestyén A, Gugolya Z, Végh BM, Vonderviszt F. Interaction of FliS flagellar chaperone with flagellin. FEBS Lett. 2006;580:3916–20.
Nambu T, Kutsukake K. The Salmonella FlgA protein, a putativeve periplasmic chaperone essential for flagellar P ring formation. Microbiology. 2000;146:1171–8.
Liu Z, Frigaard N-U, Vogl K, Iino T, Ohkuma M, Overmann J, et al. Complete genome of ignavibacterium album, a metabolically versatile, flagellated, facultative anaerobe from the phylum chlorobi. Front Microbiol. 2012;3:185.
Ferry JG. Methane from acetate. J Bacteriol. 1992;174:5489–95.
Stams AJM, Teusink B, Sousa DZ. Ecophysiology of Acetoclastic Methanogens. In Stams AJM, Sousa DZ (eds.), Handbook of Hydrocarbon and Lipid Microbiology, Vol. 2 - Biogenesis of Hydrocarbons, Springer International Publishing AG, part of Springer Nature 2018, 2019. ISBN: 978-3-319-78107-5, 1–14.
Huser BA, Wuhrmann K, Zehnder AJB. Methanothrix soehngenii gen. nov. sp. nov., a new acetotrophic non-hydrogen-oxidizing methane bacterium. Arch Microbiol. 1982;132:1–9.
Callaghan AV, Wawrik B. AnHyDeg: a curated database of anaerobic hydrocarbon degradation genes. GitHub Oklahoma. 2016.
Mayumi D, Mochimaru H, Tamaki H, Yamamoto K, Yoshioka H, Suzuki Y, et al. Methane production from coal by a single methanogen. Science. 2016;354:222–5.
Botheju D, Samarakoon G, Chen C, Bakke R. An experimental study on the effects of oxygen in bio-gasification; Part 1. Proceedings of the International Conference on Renewable Energies and Power Quality (ICREPQ 10). Granada, Spain: icrepq.com; 2010.
Angel R, Claus P, Conrad R. Methanogenic archaea are globally ubiquitous in aerated soils and become active under wet anoxic conditions. ISME J. 2012;6:847–62.
Angel R, Matthies D, Conrad R. Activation of methanogenesis in arid biological soil crusts despite the presence of oxygen. PLoS One. 2011;6:e20453.
Krzycki JA, Zeikus JG. Characterization and purification of carbon monoxide dehydrogenase from Methanosarcina barkeri. J Bacteriol. 1984;158:231–7.
An D, Caffrey SM, Soh J, Agrawal A, Brown D, Budwill K, et al. Metagenomics of hydrocarbon resource environments indicates aerobic taxa and genes to be unexpectedly common. Environ Sci Technol. 2013;47:10708–17.
Barry KP, Taylor EA. Characterizing the promiscuity of LigAB, a lignin catabolite degrading extradiol dioxygenase from Sphingomonas paucimobilis SYK-6. Biochemistry. 2013;52:6724–36.
Galushko AS, Schink B. Oxidation of acetate through reactions of the citric acid cycle by Geobacter sulfurreducens in pure culture and in syntrophic coculture. Arch Microbiol. 2000;174:314–21.
Mahadevan R, Bond DR, Butler JE, Esteve-Nuñez A, Coppi MV, Palsson BO, et al. Characterization of metabolism in the Fe(III)-reducing organism Geobacter sulfurreducens by constraint-based modeling. Appl Environ Microbiol. 2006;72:1558–68.
Neuer G, Bothe H. The pyruvate: Ferredoxin oxidoreductase in heterocysts of the cyanobacteriuim Anabaena cylindrica. Biochimica et Biophysica Acta (BBA) - Gen Subj. 1982;716:358–65.
Furdui C, Ragsdale SW. The role of pyruvate ferredoxin oxidoreductase in pyruvate synthesis during autotrophic growth by the Wood-Ljungdahl pathway. J Biol Chem. 2000;275:28494–9.
Enjalbert B, Millard P, Dinclaux M, Portais J-C, Létisse F. Acetate fluxes in Escherichia coli are determined by the thermodynamic control of the Pta-AckA pathway. Sci Rep. 2017;7:42135.
Coskun ÖK, Pichler M, Vargas S, Gilder S, Orsi WD. Linking uncultivated microbial populations and benthic carbon turnover by using quantitative stable Isotope Probing. Appl Environ Microbiol. 2018;84:e01083–18.
Nolla-Ardèvol V, Peces M, Strous M, Tegetmeyer HE. Metagenome from a Spirulina digesting biogas reactor: analysis via binning of contigs and classification of short reads. BMC Microbiol. 2015;15:277.
Li Y, Liu M, Che X, Li C, Liang D, Zhou H, et al. Biochar stimulates growth of novel species capable of direct interspecies electron transfer in anaerobic digestion via ethanol-type fermentation. Environ Res. 2020;189:109983.
Li H-Y, Wang H, Wang H-T, Xin P-Y, Xu X-H, Ma Y, et al. The chemodiversity of paddy soil dissolved organic matter correlates with microbial community at continental scales. Microbiome. 2018;6:187.
Zeng Q, Huang L, Ma J, Zhu Z, He C, Shi Q, et al. Bio-reduction of ferrihydrite-montmorillonite-organic matter complexes: effect of montmorillonite and fate of organic matter. Geochim Cosmochim Acta. 2020;276:327–44.
Maier RM. Chapter 16 - Biogeochemical Cycling. In: Pepper IL, Gerba CP, Gentry TJ (eds). Environmental Microbiology (Third Edition). 2015. Academic Press, San Diego, pp. 339–73.
Stepanauskas R, Fergusson EA, Brown J, Poulton NJ, Tupper B, Labonté JM, et al. Improved genome recovery and integrated cell-size analyses of individual uncultured microbial cells and viral particles. Nat Commun. 2017;8:84.
Eren AM, Vineis JH, Morrison HG, Sogin ML. A filtering method to generate high quality short reads using illumina paired-end technology. PLoS One. 2013;8:e66643.
Li D, Liu C-M, Luo R, Sadakane K, Lam T-W. MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015;31:1674–6.
Eren AM, Esen ÖC, Quince C, Vineis JH, Morrison HG, Sogin ML, et al. Anvi’o: an advanced analysis and visualization platform for ’omics data. PeerJ. 2015;3:e1319.
Pritchard L, Glover RH, Humphris S, Elphinstone JG, Toth IK. Genomics and taxonomy in diagnostics for food security: soft-rotting enterobacterial plant pathogens. Anal Methods. 2016;8:12–24.
Langmead B. Aligning short sequencing reads with Bowtie. Curr Protoc Bioinforma. 2010; Chapter 11: Unit 11.7.
Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–7.
Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–42.
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–10.
Pruitt KD, Tatusova T, Maglott DR. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–4.
Katoh K, Misawa K, Kuma K-I, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–66.
Criscuolo A, Gribaldo S. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol Biol. 2010;10:210.
Hyatt D, Chen G-L, Locascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinforma. 2010;11:119.
Kanehisa M, Goto S, Kawashima S, Nakaya A. The KEGG databases at GenomeNet. Nucleic Acids Res. 2002;30:42–6.
Zhou Z, Tran P, Liu Y, Kieft K, Anantharaman K. METABOLIC: A scalable high-throughput metabolic and biogeochemical functional trait profiler based on microbial genomes. Cold Spring Harbor Laboratory. 2019, 761643.
Duarte M, Jauregui R, Vilchez-Vargas R, Junca H, Pieper DH. AromaDeg, a novel database for phylogenomics of aerobic bacterial degradation of aromatics. Database. 2014;2014:bau118.
Molofsky LJ, Richardson SD, Gorody AW, Baldassare F, Black JA, McHugh TE, et al. Effect of different sampling methodologies on measured methane concentrations in groundwater samples. Ground Water. 2016;54:669–80.
Orem W, Tatu C, Varonka M, Lerch H, Bates A, Engle M, et al. Organic substances in produced and formation water from unconventional natural gas extraction in coal and shale. Int J Coal Geol. 2014;126:20–31.
Acknowledgements
BONCAT-FACS and metagenomic sequencing were conducted under CSP503725 by the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, which is supported under Contract No. DE-AC02-05CH11231. The authors (LJM, HJS, MWF) appreciate support from ENIGMA- Ecosystems and Networks Integrated with Genes and Molecular Assemblies (http://enigma.lbl.gov), a Science Focus Area Program at Lawrence Berkeley National Laboratory. We would like to thank Dr. Steven Singer at Lawrence Berkeley National Laboratory for sharing information regarding the Chlorobi NICIL-2 genome for comparison with Chlorobi PRB. We appreciate assistance in field work from Dr. Katie Davis and George Platt, and we are grateful to Dr. Jennifer MacIntosh and Dr. Daniel Ritter for geochemical analyses and discussion. We also acknowledge the USGS Energy Resources Program (Alicia Lindauer, Program Coordinator), the USGS National Innovation Center (Jonathan Stock, Director) and Montana Emergent Technologies for assistance in the field and SES development. Disclaimer: Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the U.S. Government.
Author information
Authors and Affiliations
Contributions
HJS, HDS, EPB, LJM, and MWF designed the study. EPB, HJS, and HDS performed field sampling. HJS and HDS conducted laboratory experiments. RRM and DG performed cell sorting and sequencing. LJM, HDS, EPB, HJS, and MWF analyzed and interpreted the data. LJM, HJS, and EPB wrote the paper. All authors reviewed and edited the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
McKay, L.J., Smith, H.J., Barnhart, E.P. et al. Activity-based, genome-resolved metagenomics uncovers key populations and pathways involved in subsurface conversions of coal to methane. ISME J 16, 915–926 (2022). https://doi.org/10.1038/s41396-021-01139-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41396-021-01139-x
This article is cited by
-
Using click chemistry to study microbial ecology and evolution
ISME Communications (2023)
-
Metagenomic data from surface seawater of the east coast of South Korea
Scientific Data (2023)
-
Functional and molecular approaches for studying and controlling microbial communities in anaerobic digestion of organic waste: a review
Reviews in Environmental Science and Bio/Technology (2023)
-
Subsurface hydrocarbon degradation strategies in low- and high-sulfate coal seam communities identified with activity-based metagenomics
npj Biofilms and Microbiomes (2022)