Abstract
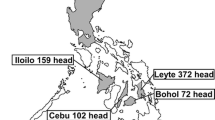
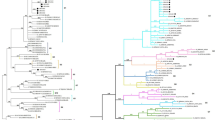
The pX genetic region of bovine leukemia virus (BLV) includes four genes with overlapping reading frames that code for the Tax, Rex, R3, and G4 proteins. These proteins are involved in the regulation of transcriptional and post-transcriptional viral expression, as well as having oncogenic potential. Our goal was to investigate the pathogenicity of the pX region of BLV genotype 1 in terms of lymphocytosis, lymphomas, and proviral DNA load. We screened 724 serological samples from mixed-age Holstein Friesian cattle from six states in Mexico. Peripheral blood leukocytes (PBLs) were isolated from whole blood with anticoagulant, and genomic DNA was extracted from the PBLs using a commercial kit. Then, a set of primers that hybridize in conserved regions of the BLV pX region were used, which allowed for PCR standardization to detect proviral DNA in infected cells. Positive amplicons were sequenced using the Sanger method, resulting in 1156-nucleotide-long final sequences that included the four pX region genes. The experimental group consisted of 30 animals. Twelve of these had lymphocytosis, six had lymphoma, and 12 were apparently healthy cattle without any signs of lymphocytosis or lymphoma. The presence of lymphoma was detected in six bovine tumor tissues using histopathology, and the presence of BLV was detected by in situ hybridization. Phylogenetic analysis demonstrated that the 30 sequences were associated with genotype 1, and the genetic distance between the sequences ranged from 0.2% to 2.09%. We identified two sequences in the G4 gene: one with a three-nucleotide deletion resulting in the loss of a leucine (AGU_7488L, in a cow with lymphocytosis), and one with a nine-nucleotide deletion resulting in the loss of leucine, proline, and leucine (AGU_18A, in a cow without lymphocytosis). Analysis of the PX region indicated that positive selection had occurred in the G4, rex, and R3 genes, and we found no difference in proviral DNA load between the studied groups. We were unable to establish an association between variations in the pX region and the development of lymphocytosis, lymphoma, asymptomatic status, or proviral DNA load in BLV-infected cattle.



Similar content being viewed by others
References
Bai L, Takeshima S, Isogai E et al (2015) Novel CD8+ cytotoxic T cell epitopes in bovine leukemia virus with cattle. Vaccine 33:7194–7202. https://doi.org/10.1016/j.vaccine.2015.10.128
Felmer R, Zúñiga J, Recabal M, Chávez R (2006) Diagnóstico y tipificación del virus de la leucosis bovina mediante una prueba de PCR-RFLP a partir de ADN extraído desde células somáticas de la leche. Archivos de medicina veterinaria. https://doi.org/10.4067/S0301-732X2006000300009
Wu MC, Shanks RD, Lewin HA (1989) Milk and fat production in dairy cattle influenced by advanced subclinical bovine leukemia virus infection. Proc Natl Acad Sci 86:993–996. https://doi.org/10.1073/pnas.86.3.993
Miller JM, Van der Maaten MJ (1982) Bovine leukosis—its importance to the dairy industry in the United States. J Dairy Sci 65:2194–2203. https://doi.org/10.3168/jds.S0022-0302(82)82482-X
Selim A, Marawan MA, Ali A-F et al (2020) Seroprevalence of bovine leukemia virus in cattle, buffalo, and camel in Egypt. Trop Anim Health Prod 52:1207–1210. https://doi.org/10.1007/s11250-019-02105-8
Gillet N, Florins A, Boxus M et al (2007) Mechanisms of leukemogenesis induced by bovine leukemia virus: prospects for novel anti-retroviral therapies in human. Retrovirology 4:18. https://doi.org/10.1186/1742-4690-4-18
El HH, Nasr R, Kfoury Y et al (2012) Animal models on HTLV-1 and related viruses: what did we learn? Front Microbiol. https://doi.org/10.3389/fmicb.2012.00333
Maezawa M, Inokuma H (2020) Analysis of bovine leukemia virus integration sites in cattle under 3 years old with enzootic bovine leukosis. Arch Virol 165:179–183. https://doi.org/10.1007/s00705-019-04431-6
Aida Y, Murakami H, Takahashi M, Takeshima S-N (2013) Mechanisms of pathogenesis induced by bovine leukemia virus as a model for human T-cell leukemia virus. Front Microbiol 4:328. https://doi.org/10.3389/fmicb.2013.00328
Krupovic M, Blomberg J, Coffin JM et al (2018) Ortervirales: new virus order unifying five families of reverse-transcribing viruses. J Virol. https://doi.org/10.1128/JVI.00515-18
Pierard V, Guiguen A, Colin L et al (2010) DNA cytosine methylation in the bovine leukemia virus promoter is associated with latency in a lymphoma-derived B-cell line: potential involvement of direct inhibition of camp-responsive element (CRE)-binding protein/cre modulator/activation transcription. J Biol Chem 285:19434–19449. https://doi.org/10.1074/jbc.M110.107607
Polat M, Takeshima S, Aida Y (2017) Epidemiology and genetic diversity of bovine leukemia virus. Virol J 14:209. https://doi.org/10.1186/s12985-017-0876-4
Sagata N, Yasunaga T, Ohishi K et al (1984) Comparison of the entire genomes of bovine leukemia virus and human T-cell leukemia virus and characterization of their unidentified open reading frames. EMBO J 3:3231–3237. https://doi.org/10.1126/science.557842
Felber BK, Derse D, Athanassopoulos A, Campbell MPG (1989) Cross-activation of the Rex proteins of HTLV-I and BLV and of the Rev protein of HIV-1 and nonreciprocal interactions with their RNA responsive elements. New Biol 1:318–328
Pozzatti R, Vogel J, Jay G (1990) The human T-lymphotropic virus type I tax gene can cooperate with the ras oncogene to induce neoplastic transformation of cells. Mol Cell Biol 10:413–417. https://doi.org/10.1128/MCB.10.1.413
Willems L, Heremans H, Chen G et al (1990) Cooperation between bovine leukaemia virus transactivator protein and Ha-ras oncogene product in cellular transformation. EMBO J 9:1577–1581
Florins A, Gillet N, Boxus M et al (2007) Even attenuated bovine leukemia virus proviruses can be pathogenic in sheep. J Virol 81:10195–10200. https://doi.org/10.1128/JVI.01058-07
Pluta A, Willems L, Douville RN, Kuźmak J (2020) Effects of naturally occurring mutations in bovine leukemia virus 5′-LTR and tax gene on viral transcriptional activity. Pathogens 9:836. https://doi.org/10.3390/pathogens9100836
Cerón Téllez F, González Méndez AS, Tórtora Pérez JL et al (2020) Lack of association between amino acid sequences of the bovine leukemia virus envelope and varying stages of infection in dairy cattle. Virus Res. https://doi.org/10.1016/j.virusres.2020.197866
Heinecke N, Tórtora J, Martínez HA et al (2017) Detection and genotyping of bovine leukemia virus in Mexican cattle. Arch Virol. https://doi.org/10.1007/s00705-017-3477-z
Mendiola WPS, Tórtora JL, Martínez HA et al (2019) Genotyping based on the LTR region of small ruminant lentiviruses from naturally infected sheep and goats from Mexico. Biomed Res Int. https://doi.org/10.1155/2019/4279573
Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754–755. https://doi.org/10.1093/bioinformatics/17.8.754
Kryazhimskiy S, Plotkin JB (2008) The population genetics of dN/dS. PLoS Genet. https://doi.org/10.1371/journal.pgen.1000304
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Tolle A, Jahnke H-D, Hasse G (2010) Zur Diagnostik der Rinderleukose und ihrer Bekämpfung in Südniedersachsen. Zentralbl Veterinarmed B 12:435–443. https://doi.org/10.1111/j.1439-0450.1965.tb01408.x
Moratorio G, Fischer S, Bianchi S et al (2013) A detailed molecular analysis of complete Bovine Leukemia Virus genomes isolated from B-cell lymphosarcomas. Vet Res 44:19. https://doi.org/10.1186/1297-9716-44-19
Lefèbvre L, Ciminale V, Vanderplasschen A et al (2002) Subcellular localization of the bovine proteins subcellular localization of the bovine leukemia virus R3 and G4 accessory proteins. J Virol 76:7843–7854. https://doi.org/10.1128/JVI.76.15.7843
Alexandersen S, Carpenter S, Christensen J et al (1993) Identification of alternatively spliced mRNAs encoding potential new regulatory proteins in cattle infected with bovine leukemia virus. J Virol 67:39–52. https://doi.org/10.1128/JVI.67.1.39-52.1993
Lefèbvre L, Vanderplasschen A, Ciminale V et al (2002) Oncoviral bovine leukemia virus G4 and human T-cell leukemia virus type 1 p13II accessory proteins interact with farnesyl pyrophosphate synthetase. J Virol 76:1400–1414. https://doi.org/10.1128/JVI.76.3.1400-1414.2002
Murakami H, Uchiyama J, Nikaido S et al (2016) Inefficient viral replication of bovine leukemia virus induced by spontaneous deletion mutation in the G4 gene. J Gen Virol 97:2753–2762. https://doi.org/10.1099/jgv.0.000583
Choi E-A, Hope TJ (2005) Mutational analysis of bovine leukemia virus rex: identification of a dominant-negative inhibitor. J Virol 79:7172–7181. https://doi.org/10.1128/JVI.79.11.7172-7181.2005
Kim FJ, Beeche AA, Hunter JJ et al (1996) Characterization of the nuclear export signal of human T-cell lymphotropic virus type 1 Rex reveals that nuclear export is mediated by position-variable hydrophobic interactions. Mol Cell Biol 16:5147–5155. https://doi.org/10.1128/MCB.16.9.5147
Zapp ML, Hope TJ, Parslow TG, Green MR (1991) Oligomerization and RNA binding domains of the type 1 human immunodeficiency virus Rev protein: a dual function for an arginine-rich binding motif. Proc Natl Acad Sci 88:7734–7738. https://doi.org/10.1073/pnas.88.17.7734
Zhao X, McGirr KM, Buehring GC (2007) Potential evolutionary influences on overlapping reading frames in the bovine leukemia virus pXBL region. Genomics 89:502–511. https://doi.org/10.1016/j.ygeno.2006.12.007
Tajima S, Aida Y (2000) The Region between amino acids 245 and 265 of the bovine leukemia virus (BLV) tax protein restricts transactivation not only via the BLV enhancer but also via other retrovirus enhancers. J Virol 74:10939–10949. https://doi.org/10.1128/JVI.74.23.10939-10949.2000
Van Den Broeke A, Bagnis C, Ciesiolka M et al (1999) In vivo rescue of a silent tax-deficient bovine leukemia virus from a tumor-derived ovine B-cell line by recombination with a retrovirally transduced wild-type taxgene. J Virol 73:1054–1065. https://doi.org/10.1128/JVI.73.2.1054-1065.1999
Merimi M, Klener P, Szynal M et al (2007) Complete suppression of viral gene expression is associated with the onset and progression of lymphoid malignancy: observations in Bovine Leukemia Virus-infected sheep. Retrovirology 4:51. https://doi.org/10.1186/1742-4690-4-51
Panei CJ, Serena MS, Metz GE et al (2013) Analysis of the pX region of bovine leukemia virus in different clinical stages of Enzootic Bovine Leukemia in Argentine Holstein cattle. Virus Res 171:97–102. https://doi.org/10.1016/j.virusres.2012.08.001
Kazemimanesh M, Madadgar O, Steinbach F et al (2019) Detection and molecular characterization of bovine leukemia virus in various regions of Iran. J Gen Virol 100:1315–1327. https://doi.org/10.1099/jgv.0.001303
McGirr KM, Buehuring GC (2006) Tax & rex: overlapping genes of the deltaretrovirus group. Virus Genes 32:229–239. https://doi.org/10.1007/s11262-005-6907-z
Nakano Y, Aso H, Soper A et al (2017) A conflict of interest: the evolutionary arms race between mammalian APOBEC3 and lentiviral Vif. Retrovirology 14:31. https://doi.org/10.1186/s12977-017-0355-4
Andoh K, Kimura K, Nishimori A, Hatama S (2020) Development of an in situ hybridization assay using an AS1 probe for detection of bovine leukemia virus in BLV-induced lymphoma tissues. Arch Virol 165:2869–2876. https://doi.org/10.1007/s00705-020-04837-7
Khudhair YI, Hasso SA, Yaseen NY, Al-Shammari AM (2016) Serological and molecular detection of bovine leukemia virus in cattle in Iraq. Emerg Microb Infect 5:e56. https://doi.org/10.1038/emi.2016.60
Rajão DS, Heinemann MB, Reis JKP et al (2014) Effects of bovine leukemia virus infection on crossbred and purebred dairy cattle productive performance in Brazil. Semina Ciências Agrárias 35:891. https://doi.org/10.5433/1679-0359.2014v35n2p891
Bartlett PC, Sordillo LM, Byrem TM et al (2014) Options for the control of bovine leukemia virus in dairy cattle. J Am Vet Med Assoc 244:914–922. https://doi.org/10.2460/javma.244.8.914
Juliarena MA, Gutierrez SE, Ceriani C (2007) Determination of provirus load in bovine leukemia virus-infected cattle with and without lymphocytosis. Am J Vet Res 68:1220–1225. https://doi.org/10.2460/ajvr.68.11.1220
Lo C-W, Borjigin L, Saito S et al (2020) BoLA-DRB3 Polymorphism is associated with differential susceptibility to bovine leukemia virus-induced lymphoma and provirus load. Viruses 12:352. https://doi.org/10.3390/v12030352
Kobayashi T, Inagaki Y, Ohnuki N et al (2020) Increasing Bovine leukemia virus (BLV) provirus load is a risk factor for progression of Enzootic bovine leucosis: a prospective study in Japan. Prev Vet Med 178:104680. https://doi.org/10.1016/j.prevetmed.2019.04.009
Martin D, Arjona A, Soto I et al (2001) Comparative study of PCR as a direct assay and ELISA and AGID as indirect assays for the detection of bovine leukaemia virus. J Vet Med Ser B 48:97–106. https://doi.org/10.1046/j.1439-0450.2001.00424.x
Kalvatchev Z, Walder R, Garzaro D, Barrios M (2000) Detection of genetic diversity among bovine immunodeficiency virus population by single-strand conformation polymorphism analysis. Viral Immunol 13:373–381. https://doi.org/10.1089/08828240050144680
Monroy Basilio JL, Tavera FJT, De Aluja AS et al (1992) Estudio comparativo entre las pruebas de ELISA e inmunodifusión en el diagnóstico de la Leucosis Enzoótica Bovina. Vet Mexico 1993:21–25
USDA (2008) Bovine Leukosis Virus (BLV) on U.S. Dairy Operations. USDA, New York, pp 1–2
Hsieh J-C, Li C-Y, Hsu W-L, Chuang S-T (2019) Molecular epidemiological and serological studies of bovine leukemia virus in Taiwan dairy cattle. Front Vet Sci. https://doi.org/10.3389/fvets.2019.00427
Callebaut I, Vonèche V, Mager A et al (1993) Mapping of B-neutralizing and T-helper cell epitopes on the bovine leukemia virus external glycoprotein gp51. J Virol 67:5321–5327. https://doi.org/10.1128/JVI.67.9.5321-5327.1993
Mamoun RZ, Morisson M, Rebeyrotte N et al (1990) Sequence variability of bovine leukemia virus env gene and its relevance to the structure and antigenicity of the glycoproteins. J Virol 64:4180–4188
Rodriguez SM, Golemba MD, Campos RH et al (2009) Bovine leukemia virus can be classified into seven genotypes: evidence for the existence of two novel clades. J Gen Virol 90:2788–2797. https://doi.org/10.1099/vir.0.011791-0
Balić D, Lojkić I, Periškić M et al (2012) Identification of a new genotype of bovine leukemia virus. Arch Virol 157:1281–1290. https://doi.org/10.1007/s00705-012-1300-4
Polat M, Takeshima S, Hosomichi K et al (2016) A new genotype of bovine leukemia virus in South America identified by NGS-based whole genome sequencing and molecular evolutionary genetic analysis. Retrovirology 13:4. https://doi.org/10.1186/s12977-016-0239-z
Lee EJ, Kim EJ, Ratthanophart J et al (2016) Molecular epidemiological and serological studies of bovine leukemia virus (BLV) infection in Thailand cattle. Infect Genet Evol 41:245–254. https://doi.org/10.1016/j.meegid.2016.04.010
OIE (2012) Enzootic bovine leukosis. OIE terrestrial manual. OIE, Paris, pp 721–732
Acknowledgements
We thank the ranchers who kindly provided samples, as well as the staff of the virology, genetics, and molecular biology laboratory of FES Cuautitlan UNAM. Neli Montero was a student in the MSc program: Programa en Ciencias de la Producción y de la Salud Animal, UNAM, and supported by a CONACyT grant scholarship.
Funding
This study was funded by FONSEC SAGARPA-CONACyT project 2017-6-292826 and by PIAPI2041. FESC. UNAM “Estudio de la respuesta inmune y genética retroviral”.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no financial or personal interests that could influence or bias the content of this article. The authors declare that they have no competing interests. All authors have seen and approved the manuscript.
Additional information
Handling Editor: William G Dundon.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Montero Machuca, N., Tórtora Pérez, J.L., González Méndez, A.S. et al. Genetic analysis of the pX region of bovine leukemia virus genotype 1 in Holstein Friesian cattle with different stages of infection. Arch Virol 167, 45–56 (2022). https://doi.org/10.1007/s00705-021-05252-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-021-05252-2