Abstract
The seed is an inevitable element for agricultural and industrial production. The non-destructive paddy seed variety identification is essential to assure paddy purity and quality. This research is aimed at developing a computer vision-based system to identify paddy varieties using multiple heterogeneous features, exploiting textural, external, and physical properties. We captured the paddy seed images without any fixed setup to make the system user friendly at both industry and farmer levels, which can lead to illumination problems in the images. To overcome this problem, we introduced a modified histogram oriented gradient (T20-HOG) feature that can describe the illumination, scale, and rotational variations of a paddy image. We also utilized the existing Haralick and traditional features and the dimensionality of the features is reduced by the Lasso feature selection technique. The selected features are used to train the feed-forward neural network (FNN) to predict the paddy variety. The experiments conducted on two different datasets: BDRICE, and VNRICE. Results of our method are shown in terms of four standard evaluation metrics, namely, accuracy, precision, recall, and F_1 score, and achieved 99.28%, 98.64%, 98.48%, and 98.56% score, respectively. We also compared our system efficiency with existing studies. The experimental results demonstrate that our proposed features are effective to identify paddy variety and achieved a new state-of-the-art performance. And we also observed that our newly proposed T20-HOG features have a major impact on overall system performance.
Similar content being viewed by others
Introduction
Globally, paddy is the second most important cereal crop in terms of production area. It is also the staple food for nearly half of the world’s people.Footnote 1 Agriculture is the largest employment sector in Bangladesh, and it is estimated that 71.9% of the total population lives in rural areas, while 47% of the labor forces are engaged in agriculture.Footnote 2
Bangladesh will require about 44.6 million tons of paddy in the year 2050 [31]. During this time, the total paddy cultivation area will also be shrunk to 10.28 million hectares. Therefore, paddy yield needs to be increased by nearly 1.3 tons per hector. There are many factors involved in increasing crop yield. Among them, seed quality is one of the vital factors. Nobody can expect better crop yield in any circumstances without genetically pure (e.g., a type of paddy seed variety that does not mix with other types of paddy seed varieties) quality seed [33, 37]. It has experimentally shown that using good quality seed paddy yield could be increased to 15%–20% [26]. However, the mixture of different paddy varieties causes adulteration in the rice market, which reduces the economic value of rice and also the trust between buyer and seller [4, 54]. Paddy seed varieties are almost similar from outside, and it is tough to differentiate manually [18]. If the farmer cultivates the wrong type of varieties by assuming their intended variety of paddy, the overall productivity might be decreased. Because, different varieties need different types of soil, methods of cultivation, as well as the duration of maturity [57].
There exist some laboratories for seed purity testing, where seed varieties are identified by human experts which require a notable amount of time and cost. To overcome these problems, different researchers have given their considerable attention to develop automatic seed grading system using classical image processing and computer vision techniques [8, 10, 23, 24, 29, 36, 38, 45, 50, 52, 58, 59].
Different researchers [1, 13, 28, 34, 40, 44, 59] concentrated on paddy grain classification from a single kernel where they used heterogeneous features including geometrical, morphological, statistical, color, and textural features. Geometric features were utilized by several studies [1, 12, 44, 47] with the combinations of other features. Because, only geometric features are not efficient for achieving high accuracy. A morphological feature can be a good representative to achieve the high accuracy [12, 15, 44, 51, 55, 59]. However, the morphological features may provide the wrong direction when the dataset contains images of different resolution. Many researchers used several statistical properties of an image [12, 22], but these are sensitive to scale and rotational variations. Color can also be a good feature for paddy identification, because it varies over different paddy varieties. Researchers tried to use color features and conducted several studies [1, 44, 46, 51]. They also mentioned that this feature is very much sensitive to the room environment as well as lighting conditions. To overcome this problem, scientists proposed different textural features [1, 3, 12, 28, 46, 47, 51] including Haralick [3, 6], HOG [19, 41], GIST [28], and SIFT [28]. Most of the textural feature generates a long vector which leads to increase system time complexity.
Another important part of paddy classification is to choose a perfect machine learning model, because an appropriate algorithm can provide a high performance. Different researchers applied artificial neural network (ANN) [3, 6, 12, 46, 59], support vector machine (SVM) [47, 59], and deep learning [27] techniques for paddy variety identification. From the literature review of the mentioned studies, it is easy to say that the ANN outperformed other methods. However, the performance of classification techniques can be increased by reducing the time using different feature selection methods [16, 19, 20, 61].
From the above literature review, we understood that many research works were conducted to identify paddy variety using machine learning and computer vision approaches. Most of the research works achieved a good performance; however, they used their self prepared dataset rather than validating with publicly available datasets, which are varying by size, resolution, types, environment, and lighting conditions. Again, most of the dataset was prepared to target a single kernel, while a few studies used multiple kernels on a single image. Interestingly, we observed that most of the high performing studies used their self dataset. We also observed that the textural feature generates a large vector. Therefore, an efficient modification of textural features is needed to reduce the time complexity.
In this research, we aim to develop a computer vision-based paddy variety identification system by exploiting heterogeneous features from paddy seeds. This system will be very useful for both industries and end-user levels. At the industry level, this system can be used at the packaging time using a conveyor belt, so that different categories of seeds are not mixed together. However, at the farmer level, the farmer can also verify their seed using our model. We also aim to develop a paddy dataset of large volume and compare our method with standard datasets. In this research, we used two datasets: BDRICE and VNRICE. BDRICE is prepared by us, which is a multiple kernel image dataset where the images were captured using a smartphone without a fixed setup. VNRICE is a standard paddy dataset of a single kernel, several research works have been conducted on this dataset [19, 27, 41]. We extracted heterogeneous features and proposed a new feature set. After that, a feature selection technique is used to reduce the dimensionality problem. Then, we trained different machine learning algorithms and select the best model according to different evaluation metrics.
This work is organized as follows. First, we present the details of our proposed method to identify paddy seed variety in “Methodology”. In “Experiments and evaluation”, we discussed experimental results and evaluation. Finally, we summarize our work and draw some conclusions and future directions “Conclusion”.
Methodology
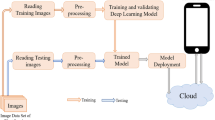
This section presents our proposed method for paddy variety identification. Our method broadly has five steps, namely, i. image acquisition, ii. image processing, iii. feature extraction, iv. feature selection, and v. training model. The block diagram of our proposed system is shown in Fig. 1.
Sample images from two datasets: BDRICE and VNRICE. Here, a–g represent BDRICE which is our prepared dataset. Where a–d is a multiple kernel paddy image dataset of four varieties BRRI 11, BRRI 28, BRRI 29, and BRRI 81 are represented sequentially. e–g represents the final single kernel paddy dataset segmented from a–d by applying image preprocessing step. And (h–m) VNRICE a reference dataset, where BC-15, Huong Thom 1, Nep-87, Q-5, Thien Uu-8, and Xi-23 varieties are presented sequentially
Image acquisition The experiment was carried out collecting paddy from the Bangladesh Rice Research Institute (BRRI). Different varieties of paddy, namely, BRRI Dhan 11, BRRI Dhan 28, BRRI Dhan 29, and BRRI Dhan 81, were collected and cleaned to free from impurities and foreign matters. Pure paddy seed was selected through purity test according to ISTA, 1999, and by separating other seeds, i.e., other crop seeds, other paddy variety seeds from seed lot. Then, purified paddy of each variety have taken to the experimental area for sampling individually. In our experimental lab, cleaned paddy were placed on black cloth with the paddy not touching each other. After that, a smartphone (rear camera 8MP, 1.4GHz quad-core Qualcomm Snapdragon 425 processor) was used to acquire the images at an average height of 15–20 cm where the lighting condition was not fixed. The images were saved in JPG format. The sample images are shown in Fig. 2.
Image preprocessing We applied different image processing techniques on paddy seed images such as binarization, individual seed segmentation, noise removing, overlapped seed segmentation, and hole filling. In the first step, we converted the RGB images into gray images and then binary images using the function rgb2gray and im2bw in MATLAB 2020a. In the next step, we applied MATLAB regionprops function with 8-connectivity for segmenting the paddy regions. After that, we analyzed all the regions to remove the noisy areas which were less than 100 pixels using bwareaopen method. In the final step, we applied watershed and imfill methods to split the overlapped paddy seed regions, and to fill the holes, respectively, if there any.
Feature extraction In computer vision and machine learning algorithms, feature extraction plays a vital role in classification. Features are the essential parameters that can illustrate an image or image portions. A feature will be appropriate if the representative value remarkably varies over different classes or groups. We extracted various features for each paddy image, e.g., traditional, Haralick, and T20-HOG for training the FNN model.
-
(A)
Traditional features The summary of external and physical features is illustrated in Table 1. In nature, the maximum of the paddy grain seed color varies from yellow–green to yellow, and yellow to yellow-harden. For some of the varieties, colors vary from light black to deep black. Hence, the surface color feature can be used for describing individual paddy [5]. Again, color is invariant with respect to scale, translation, and rotation, so it can be an important feature to describe paddy seed [4, 34, 42]. Some seeds are smaller in length compared to others, while some are wider as well. Based on the length and width, paddy varieties are categorized into three types long, medium, and short. Therefore, the length and width of the paddy can be a distinctive descriptor [34, 35]. Flatten and rounded paddy shapes can be another distinguishing point. The total surrounding border describes an object’s elongation, which also describes the seed is broken or not; hence, perimeter can be an important feature [7, 13, 34, 35]. The area expresses the amount of surface within the border. A maximum node possible polygon in a region describes all angular tuning points or corners in an object, which is represented as solidity in paddy. Hence, area and solidity are another shape descriptor for paddy variety identification [34]. Different researchers used these features in paddy variety identification [44, 51].
-
(B)
Haralick features Texture means patterns or variations in an image surface. It is defined by the spatial distribution of neighborhood pixels of a surface [60]. It can describe the object of different scales, rotation, and resolution [25]. The gray-level co-occurrence matrix (GLCM) describes how frequently the different patterns exist including energy, contrast, correlation, roughness, and entropy of an image [4]. Fourteen different Haralic [13] textural features can narrate a paddy seed and differentiate from other varieties. Let, the dimension of any co-occurrence matrix is \(N_g \times N_g\). The frequency of the gray-level values is \(N_g\), and the matrix spatial distributions of neighborhood pixel are \(P_d\),. And \(P_d(i,j)\) indicates (i, j)th elements of GLCM matrix. The details are presented in Table 2.
-
(C)
T20-HOG HOG features are used for different types of object detection tasks including pedestrian [11, 14], human [49], crop pest [17], and palm tree [56] detection. Basically, these features compute the gradients’ orientations of pixels in an object image. Then, it counts the frequency and creates a vector that is treated as a global descriptor of an object. These gradients can describe the illumination [43],Footnote 3 scale [21] and rotational variations [2] of an object. Practically, the size of the vector is large, where a lot of values are repeated. If we can shrink that large vector, it will reduce the dimensionality problem. To reduce the size of the vector by selecting the important features, we analyzed it in different ways, so that it can play a significant role to identify the object as well as reducing the time complexity. In this process, we first extracted the HOG features using the function extracthogfeatures in MATLAB. Usually, this function returns a vector of different dimensions for different sizes of input images, and the vector contains an enormous repetition of values. To solve the problems, we proposed a new Algorithm 1 called T20-HOG which can reduce the dimensionality problem as well as repetitions of the values.

Lasso feature selection Feature selection algorithms are used to find the relevant features from a feature set. It tells us which features to consider, which ones to discard, and which ones have a negative impact on overall performance [16, 20, 61]. Lasso is one of the feature selection methods which is widely used for classification problems. Lasso stands for least absolute shrinkage and selection operator. This technique works based on linear regression with L1 regularizationFootnote 4 which is measured by
where \(x^{(i)}\) is the vector of actual value, \(y^{(i)}\) is the vector of predicted value, m indicates the number of observation, \(\lambda \) is the regularization parameter, \(\theta _{j}\) is the gradient descent, and the cost function is
where
We provide \(X \times Y\) dimensions feature matrix and \(X \times 1\) dimensional level vector to the Lasso system, where X indicates the number of initial features and Y indicates the number of paddy seed. The Lasso function computes feature influence for every feature using the above formulas. Finally, it returns an Z vector which contains the ranking of features, where \(Z \le X\). These selected \( Z \times Y\) matrix and \(X \times 1\) dimensional level vector are passed to the FNN model. By applying this Lasso feature selection technique, we obtained the following important features from the three different groups of features depicted in Table 3.
Feed forward neural network model (FNN) After applying the Lasso feature selection technique, the selected features are feed into the FNN model to identify the paddy variety. FNN offers exciting possibilities to perform machine learning and the prediction that is utilized and performing in different areas of the real world including agriculture [9, 32, 39, 48, 53]. It can detect the suitable pattern from input or training data. FNN is a special type of ANN, where information moves only in the forward direction, from input to hidden nodes, and then hidden to output nodes. There is no cyclic information flow in FNN. In our proposed system, we used 3 hidden layers, each containing 30 nodes and one node in the output layer. The selected features are considered as input in FNN. The training process is described below.
Let \(\mathbf {X}\) be the input vector consists of all the selected features and \(\mathbf {W}_{h}\) and \(\mathbf {W}_{o}\) be the weight vector for the hidden layer and the output layer, respectively. The input vector is used by the hidden layer and the output of all neurons in the hidden layer are computed as the following tansig activation function:
where \(V_{h}\) denotes the result calculated by \(\mathbf {X}\) and \(\mathbf {W}_h\). The output of the hidden layer \(\mathbf {Y}_h\) is feed in the output layer as input vector. In this layer, we applied sigmoid activation function for the final outcome as follows:
The difference between target \(\mathbf {Y}\) and predicted \(\hat{\mathbf {Y}}\) output is termed as error and calculated based on the square of difference between these two values using the following equation:
where m is the total number of seeds to train. To build an optimal model, the goal is to minimize the error amount l.
We divided our dataset into two sets: one is for training (75%) and the other is for testing (25%), using random selection. The training set is used to train the network and the testing set is used for computing the accuracy. We tested the trained FNN with the real world testing samples. This network is expected to identify the variety of the seed for a given input set.
Testing the model Our system segmented each paddy from captured paddy images using different image processing algorithms. Later, the features of that paddy are extracted. And all the features of the paddy are given to the trained model. It tells us the name of the paddy varieties. After that, we marked the paddy image by the obtained variety name. We also developed a software interface based on the image testing system that takes an image with multiple paddies. Our system identified each of the paddy varieties and marked them with a square boundary with their resultant variety name. The system is very simple to use by farmers. The sample output is shown in Fig. 3.
Experiments and evaluation
This section presents the dataset collection, experimental setup, experimental results, findings, and discussions. We will also discuss the efficiency of different features with different machine learning models.
Dataset and experimental configurations
In our experiment, we used two different dataset: BDRICE (prepared by us) and VNRICE [19, 27, 41]. The BDRICE consists of 60,800 paddy seed images of four different varieties. The VNRICE [19, 27, 41] is comprised with six paddy varieties. The description of both datasets is given in Table 4. We used 75% data for training the model and 25% for testing purposes.
Experimental setup To evaluate the proposed model’s effectiveness, we conducted experiments on eight different settings. The names and the description of the settings and their corresponding results are described in Table 5.
We also evaluated our proposed system in terms of robustness by conducting experiments on two different settings. In the first setting, we have taken different combinations of paddy varieties. In the second setting, we have changed the ratio of training and testing images. We conducted all the above experiments on the BDRICE dataset, and the best result is applied on the VNRICE dataset to evaluate our system performance with other models.
Machine learning model configuration In this study, we used four different ML models: FNN, SVM, K-nearest neighbor (KNN), and decision tree (DT). We configured FNN with 30 nodes, 3 hidden layer, 1 output layer, tansig activation function in hidden layer, and sigmoid activation function in output layer. In case of SVM, we used multiclass model support vector machine onevsall technique with Gaussian kernel where the learning parameters were matlab default values. Again, we used the configuration for KNN with 4-neighbors, mahalanobis distance, squaredinverse weight, and quadratic loss function. Finally, we configured DT with MaxNumSplits 4, KFold 10, optimizer bayesopt, acquisition function expected-improvement, and repartition status false.
Experimental results
To evaluate the performance of our experiments, we used a tenfold cross-validation technique with four different machine learning models: FNN, SVM, K-nearest neighbor (KNN), and decision tree (DT). The performance was evaluated utilizing four different evaluation metrics: accuracy, precision, recall, and F1_score. Let, we have five classes in our dataset, and then, we get five values for each evaluation metric (e.g., accuracy) in each setup. For brevity, from there, we present only the best result of five values.
Our experimental results are described in Table 5. We observed that the minimum performance achieved in Trad_NM settings, and the maximum performance in Lasso_NM settings where the difference between the accuracy of these two settings is 78.75%. The performance of evaluation metrics indicates that Lasso_NM performing better than all the other techniques. This can also be identified from the results of all the machine learning model’s accuracy, where the accuracy varies from 0.05% to 4%.
We present our confusion matrix for the BDRICE dataset in Fig. 4 using the features obtained from Lasso_NM settings with the FNN model. In the confusion matrix, we use actual and predicted levels for comparison, and then, we calculate different evaluation metrics from the matrix. Finally, we select the best results among all varieties.
Performance comparison of different varieties on BDRICE dataset using Lasso_NM settings from Table 5 with FNN model. Here, we use different evaluation measures including true positive (TP), true negative (TN), false positive (FP), and false negative (FN). Also, we use different evaluation metrics including accuracy (Accr), precision (Prcn), recall, and F1_Score (F1)
Machine learning model selection We conducted experiments to determine the best machine learning model in terms of different evaluation metrics. From Table 5, we can see that the highest performance is achieved on Lasso_NM settings, and applied it to find the best machine learning model. The experimental results are shown in Fig. 5, and we achieved high performance for all the models, where the minimum accuracy is 97.66%, and the maximum one is 99.28%. In terms of different evaluation metrics, we observed that the best and lowest performance achieved on FNN, DT, where the difference of precision and F1_score is 2.58% and 1.79%, respectively.
Performance comparison of different machine learning models with features selected by Lasso Lasso_NM settings from (Table 5)
Necessity of feature selection We experimented to determine the importance of feature selection in machine learning. We used two different settings: features without filtering AF_NM and features selected by Lasso LS_NM (Table 5). From Fig. 6 we see that between these two settings, the recall value varies from 8% to 11% and the accuracy varies from 7% to 9% where LS_NM setting provides a good score in both cases. We also observe that the FNN contributes significantly in terms of accuracy, whereas KNN performs well for recall value. These results reveal that feature selection boosts performance and plays a vital role to develop an appropriate model.
Performance analysis of feature selection: all features AF_NM and features selected by Lasso LS_NM (Table 5)
Performance stability on the number of variety We conducted an experiment to visualize the performance effect of changing the number of paddy varieties. First, we obtained the accuracy by considering two varieties termed as 2V_NM which consists of all combinations of two varieties determined by \(_{4}C_{2}\) formula. It means that there are 6 subsets {(BRRI11, BRRI28),(BRRI11, BRRI29),(BRRI11, BRRI81),(BRRI28, BRRI29), (BRRI28, BRRI81), and (BRRI29, BRRI81)}. All the subsets are examined with a tenfold cross-validation technique, and average accuracy is shown in Fig. 7. Similarly, we experimented on three and four varieties termed as 3V_NM and 4V_NM, respectively. For all settings, we used the FNN model to train features. We observed that with the number of varieties, the accuracy changes slightly which implies that if we change the number of paddy varieties, it will not have much effect on the overall performance of the model.
Model robustness on training and testing ratio: We also examined the effect of changing ratio in training and testing datasets. We considered four different settings: 20:80, 40:60, 60:40, and 80:20 termed as 2BY8_NM, 4BY6_NM, 6BY4_NM, and 8BY2_NM respectively, where the numbers represents the training and testing image percentages. We conducted a tenfold cross-validation technique for all varieties using Lasso_NM features with the FNN model and the best result is depicted in Fig. 8. We observed the nominal performance difference between the setting, where the minimum accuracy is 98.76% for 2BY8_NM and maximum accuracy 99.28% for 8BY2_NM settings. To conclude, the number of paddy seeds in training and testing is not influencing remarkably which demonstrates the robustness of the proposed system.
Feature efficiency To evaluate the influences of proposed textural features over traditional features on the model building, we conducted experiments in two settings. The first one includes the traditional features described in Table 1 which is expressed as OTF_NM. In another setting, we considered the textural features of both Haralick and our newly proposed T20-HOG which is denoted by HTF_NM (Table 5). The experiment results are depicted in Fig. 9. We observed that traditional features have less importance rather than textural features and the differences between accuracy and precision on two settings are 64%, and 65%, respectively. In addition, for all evaluation metrics, HTF_NM settings provide very high performance in comparison to OTF_NM settings.
In our experiment, we have taken images of paddy in an open environment, so the resolution and depth of our pictures are different. Due to these reasons, the values of traditional features (height, width, area, perimeter, etc.) have deviated a lot. That can lead to bad results in OTF_NM setting. On the contrary, textural features address these limitations; that is why, HTF_NM (Table 5) setting provides good results.
Dominance of brand new T20-HOG: It is inferred that textural features are very powerful for our research. However, we have two different groups of textural features one is our brand new T20-HOG and the other is existing Haralick features. Therefore, we analyzed in detail to find the impact of each group of textural features. Here, we also considered two different settings. One is building and testing the model with only T20-HOG features, expressed as T20_NM (Table 5), and another setting comprises of Haralick textural features represented by HF_NM (Table 5). The result is shown in Fig. 10. From the graph, we see that the minimum result variation of the two settings is 8.29%, and the maximum is 11.66%. Overall, the performance of T20_NM settings is remarkably high, because T20-HOG features can represent the textural changes effectively and boost up the performance.
Robustness of proposed T20-HOG: We compared the performance of T20-HOG (T20_NM setting) with traditional HOG features (HOG_NM setting), shown in Table 5. The result is depicted in Fig. 11, from this figure we observed that the performance of T20-HOG outperformed the HOG in all four evaluation metrics, which indicates the robustness of our newly proposed T20-HOG features.
Moreover, we experimented by taking T40-HOG, T30-HOG, T20-HOG, and T10-HOG features from sorted HOG count values. Then, we feed these features into FNN to find the accuracy of the BDRICE dataset. Later, we applied the Lasso features selection technique on the feature set and calculated the accuracy again. The detailed result with and without feature selection, no of selected features by Lasso, and selected feature names are presented in Table 6. From the table, we can see that T20-HOG with feature selection provides the best result. Besides, the lowest performance is achieved on T40-HOG without feature selection. Hence, we can conclude that our proposed T20-HOG with lasso feature selection is highly effective for paddy classification.
Performance analysis of newly proposed T20-HOG T20_NM over traditional HOG features HOG_NM (Table 5)
Comparison with standard dataset
We also used a standard dataset VNRICE to measure our system efficiency. Several research works have been conducted on this dataset [19, 27, 41] for paddy variety identification. Duong et al. [19] applied HOG and feature selection techniques to achieve better accuracy. Nguyen et al. [41] worked on the same dataset and tried to classify paddy variety by utilizing HOG and missing value imputation technique. Hoai et al. [27] applied different deep learning techniques to determine the paddy type and they achieved the highest accuracy for DenseNet121. We compared our system accuracy with the above studies, where we used our best model comprised with Lasso_NM (Table 5) features trained with the FNN model. The comparison result is shown in Table 7. From this table, we observe that our method is superior to other studies applied on the VNRICE dataset. From Table 5, we observed that T20-HOG features achieved better performance than traditional HOG, which is why our system shows better accuracy than Nguyen et al. [41].
Comparison with recent works
To evaluate the comparative performance, we considered a few prominent recent works in paddy classification. We applied their techniques in our BDRICE dataset and compared them with our results. At first, we considered Ansari et al. [7] work where they trained an SVM model by extracting twenty features: seven color, nine morphological, and four textural features. They converted input RGB image to HSV, and then applied imfill and bwareaopen for removing small objects under 100 pixels. We applied their technique in our dataset and found accuracy 78.21% which is presented in Table 8. However, it is far less than our proposed method’s accuracy.
We also considered Javanmardi et al. [30] study where they used VGG16 convolutional neural network (CNN) architecture for feature extraction from corn seeds. They extracted corn seed features using the VGG16 CNN model and these features are feed into the ANN model. We applied their technique in our BDRICE dataset where the configurations were Image size: \(224 \,\times \, 224 \,\times \, 3\), depth: 16, optimizer: RMSprop, loss function: cross-entropy, max epochs: 100, batch size: 32, learning rate: 0.01, parameters: 138 M. The accuracy of this experiment is 99.36% which is identical to our proposed model. Finally, we can say that our method is effective and suitable for paddy seed classification.
In summary, we can say with conviction that feature selection significantly improves performance. Interclass similarity and intraclass variability are clearly visible in our features selection. The system performance is almost stable when we changed the number of paddy varieties as well as the training and testing ratio. The textural features are efficient in identifying paddy seeds’ variety. Our brand new T20-HOG feature has a notable impact on overall performance in comparison to the traditional HOG, while the combination of T20-HOG and Haralick features boost the system performance. We also applied our system to the VNRICE dataset and compared it with the existing works and observed that our system outperformed all the existing systems which indicates the robustness of our new system.
Conclusion
In this paper, we proposed a new method to identify paddy variety with the help of the FNN model trained with different heterogeneous features. The T20-HOG features were introduced in paddy seed identification for the first time and we found that it has a considerable impact on overall performance as compared to the other textural features and traditional features. We applied the feature selection techniques to find the impact of each feature in the model building that helps to reduce the dimensionality. The accuracy of experimental results on feature selection is 99.28% which concluded that our introduced features have a great impact to identify paddy variety. We also evaluated the system performance by changing the number of varieties, the number of train, and test ratios. For all experimental settings, our method showed promising performance in different evaluation metrics. The system is also applied on the VNRICE dataset for comparison with existing systems which also applied on that dataset. We observed that our accuracy on that dataset is 99.71%, whereas the accuracy of other models Duong et al. [19], Nguyen et al. [41], and Hoai et al. [27] DenseNet121 are 93.34%, 99.66% and 99.04%, respectively. From this experiment, we can say that our result is superior among all the feature and deep learning-based methods, leading to evidence of the robustness of the newly proposed system. Moreover, we compared the performance achieved by our method with some recent existing works. The accuracy of our model is 99.28% whereas the performance of Ansari et al. [7] and Javanmardi et al. [30] are 78.21% and 99.36% respectively. These comparisons reveal that our method is superior to the Ansari et al. [7] method and almost identical to Javanmardi et al. [30] VGG16 CNN method. Hence, this system can be used in both the industry and farmers’ levels efficiently and effectively.
We prepared an annotated dataset with 60800 paddy images, which can be used as a benchmark dataset in seed variety identification. However, in this study, bulky seeds, heavily overlapped or connected paddy seeds were not considered. In the future, we would like to focus on enriching the dataset by including more available paddy varieties and increasing the number of paddy images in each variety.
Notes
IRRI, 2013 Trends in global rice consumption. Rice Today, January–March 2013 Issue. (irri.org/rice/trends-in-global-rice).
World Population Review, 2015, http://worldpopulationreview.com/countries/bangladesh-population/.
References
Abbaspour-Gilandeh Y, Molaee A, Sabzi S, Nabipur N, Shamshirband S, Mosavi A (2020) A combined method of image processing and artificial neural network for the identification of 13 Iranian rice cultivars. Agronomy 10(1):117
Alshehri H, Hussain M, Aboalsamh HA, Al Zuair MA (2018) Cross-sensor fingerprint matching method based on orientation, gradient, and Gabor-Hog descriptors with score level fusion. IEEE Access 6:28951–28968
Anami BS, Malvade NN, Hanamaratti N (2016) An edge texture features based methodology for bulk paddy variety recognition. Agric Eng Int CIGR J 18(1):399–410
Anami BS, Malvade NN, Palaiah S (2019) Automated recognition and classification of adulteration levels from bulk paddy grain samples. Inf Process Agric 6(1):47–60
Anami BS, Malvade NN, Palaiah S (2020) Classification of yield affecting biotic and abiotic paddy crop stresses using field images. Inf Process Agric 7(2):272–285
Anami BS, Naveen N, Hanamaratti N (2015) Behavior of HSI color co-occurrence features in variety recognition from bulk paddy grain image samples. Int J Signal Process Image Process Pattern Recogn 8(4):19–30
Ansari N, Ratri SS, Jahan A, Ashik-E-Rabbani M, Rahman A (2021) Inspection of paddy seed varietal purity using machine vision and multivariate analysis. J Agric Food Res 3:100109
Barbedo JG, Guarienti EM, Tibola CS (2018) Detection of sprout damage in wheat kernels using NIR hyperspectral imaging. Biosyst Eng 175:124–132
Boyaci D, Erdoğan M, Yildiz F (2017) Pixel-versus object-based classification of forest and agricultural areas from multiresolution satellite images. Turk J Electr Eng Comput Sci 25(1):365–375
Butardo VM, Sreenivasulu N (2019) Improving head rice yield and milling quality: state-of-the-art and future prospects. In: Rice grain quality. Springer, New York, pp 1–18. https://doi.org/10.1007/978-1-4939-8914-0_1
Cai L, Zhu J, Zeng H, Chen J, Cai C, Ma KK (2018) Hog-assisted deep feature learning for pedestrian gender recognition. J Frankl Inst 355(4):1991–2008
Chaugule A, Mali SN (2014) Evaluation of texture and shape features for classification of four paddy varieties. J Eng 2014. https://doi.org/10.1155/2014/617263
Chaugule AA, Mali SN (2016) Identification of paddy varieties based on novel seed angle features. Comput Electron Agric 123:415–422
Chee KW, Teoh SS (2019) Pedestrian detection in visual images using combination of hog and hom features. In: 10th International Conference on robotics, vision, signal processing and power applications. Springer, New York, pp 591–597. https://doi.org/10.1007/978-981-13-6447-1_75
Chen S, Xiong J, Guo W, Bu R, Zheng Z, Chen Y, Yang Z, Lin R (2019) Colored rice quality inspection system using machine vision. J Cereal Sci 88:87–95
Chen SB, Zhang YM, Ding CH, Zhang J, Luo B (2019) Extended adaptive lasso for multi-class and multi-label feature selection. Knowl Based Syst 173:28–36
Chu H, Zhang D, Shao Y, Chang Z, Guo Y, Zhang N (2018) Using HOG descriptors and UAV for crop pest monitoring. In: 2018 Chinese Automation Congress (CAC). IEEE, pp 1516–1519
Cuevas RP, Pede VO, McKinley J, Velarde O, Demont M (2016) Rice grain quality and consumer preferences: a case study of two rural towns in the Philippines. PLoS One 11(3):e0150345
Duong HT, Hoang VT (2019) Dimensionality reduction based on feature selection for rice varieties recognition. In: 2019 4th International Conference on Information Technology (InCIT). IEEE, pp 199–202. https://doi.org/10.1109/INCIT.2019.8912121
Fonti V, Belitser E (2017) Feature selection using lasso. VU Amsterdam Research Paper in Business Analytics, pp 1–25
Giveki D, Soltanshahi MA, Montazer GA (2017) A new image feature descriptor for content based image retrieval using scale invariant feature transform and local derivative pattern. Optik 131:242–254
Golpour I, Chayjan R et al (2014) Identification and classification of bulk paddy, brown, and white rice cultivars with colour features extraction using image analysis and neural network. Czech J Food Sci 32(3):280–287
Gong Z, Cheng F, Liu Z, Yang X, Zhai B, You Z (2015) Recent developments of seeds quality inspection and grading based on machine vision. In: 2015 ASABE Annual International Meeting. American Society of Agricultural and Biological Engineers
Grillo O, Blangiforti S, Venora G (2017) Wheat landraces identification through glumes image analysis. Comput Electron Agric 141:223–231
Hall-Beyer M (2017) Practical guidelines for choosing GLCM textures to use in landscape classification tasks over a range of moderate spatial scales. Int J Remote Sens 38(5):1312–1338
Haque A, Elazegui F, Mia MT, Kamal M, Haque MM (2012) Increase in rice yield through the use of quality seeds in Bangladesh. Afr J Agric Res 7(26):3819–3827
HOANG VT, Van Hoai DP, Surinwarangkoon T, Duong HT, Meethongjan K (2020) A comparative study of rice variety classification based on deep learning and hand-crafted features. ECTI Trans Comput Inf Technol (ECTI-CIT) 14(1):1–10
Hong PTT, Hai TTT, Hoang VT, Hai V, Nguyen TT et al (2015) Comparative study on vision based rice seed varieties identification. In: 2015 Seventh International conference on knowledge and systems engineering (KSE). IEEE, pp 377–382. https://doi.org/10.1109/KSE.2015.46.
Huang M, Wang Q, Zhu Q, Qin J, Huang G (2015) Review of seed quality and safety tests using optical sensing technologies. Seed Sci Technol 43(3):337–366
Javanmardi S, Ashtiani SHM, Verbeek FJ, Martynenko A (2021) Computer-vision classification of corn seed varieties using deep convolutional neural network. J Stored Prod Res 92:101800
Kabir M, Salam M, Chowdhury A, Rahman N, Iftekharuddaula K, Rahman M, Rashid M, Dipti S, Islam A, Latif M et al (2015) Rice vision for Bangladesh: 2050 and beyond. Bangladesh Rice J 19(2):1–18
Kaur S, Girdhar A, Gill J (2018) Computer vision-based tomato grading and sorting. In: Advances in data and information sciences. Springer, pp 75–84
Kelly AF (2013) Seed production of agricultural crops. Scientific Publishers, New York
Khazaei J, Golpour I, Ahmadi Moghaddam P (2016) Evaluation of statistical and neural network architectures for the classification of paddy kernels using morphological features. Int J Food Prop 19(6):1227–1241
Liu CC, Shaw JT, Poong KY, Hong MC, Shen ML (2005) Classifying paddy rice by morphological and color features using machine vision. Cereal Chem 82(6):649–653
Mahesh S, Jayas D, Paliwal J, White N (2015) Hyperspectral imaging to classify and monitor quality of agricultural materials. J Stored Prod Res 61:17–26
McDonald MF, Copeland LO (2012) Seed production: principles and practices. Springer, New York
Mittal S, Dutta MK, Issac A (2019) Non-destructive image processing based system for assessment of rice quality and defects for classification according to inferred commercial value. Measurement 148:106969
Mushiri T, Tende L (2020) Automated grading of tomatoes using artificial intelligence: the case of zimbabwe. In: AI and Big Data’s potential for disruptive innovation. IGI Global, pp 216–239. https://doi.org/10.4018/978-1-5225-9687-5.ch008
Neelamegam P, Abirami S, Priya KV, Valantina SR (2013) Analysis of rice granules using image processing and neural network. In: 2013 IEEE Conference on Information & Communication Technologies. IEEE, pp 879–884. https://doi.org/10.1109/CICT.2013.6558219
Nguyen-Quoc H, Hoang VT (2020) Rice seed image classification based on hog descriptor with missing values imputation. Telkomnika 18(4):1897–1903
OuYang AG, Gao Rj, Sun Xd, Pan Yy, Dong Xl et al (2010) An automatic method for identifying different variety of rice seeds using machine vision technology. In: 2010 Sixth International Conference on Natural Computation, vol 1. IEEE, pp 84–88
Patel MI, Thakar VK, Shah SK (2016) Image registration of satellite images with varying illumination level using hog descriptor based surf. Procedia Comput Sci 93:382–388
Pazoki A, Farokhi F, Pazoki Z et al (2014) Classification of rice grain varieties using two artificial neural networks (MLP and neuro-fuzzy). J Anim Plant Sci 24(1):336–343
Polak A, Coutts FK, Murray P, Marshall S (2019) Use of hyperspectral imaging for cake moisture and hardness prediction. IET Image Proc 13(7):1152–1160
Rad SJM, Tab FA, Mollazade K (2011) Classification of rice varieties using optimal color and texture features and BP neural networks. In: 2011 7th Iranian Conference on machine vision and image processing. IEEE, pp 1–5. https://doi.org/10.1109/IranianMVIP.2011.6121583.
Rad SM, Tab FA, Mollazade K (2012) Application of imperialist competitive algorithm for feature selection: a case study on bulk rice classification. Int J Comput Appl 40(16):41–48
Ramesh S, Vydeki D (2019) Recognition and classification of paddy leaf diseases using optimized deep neural network with JAYA algorithm. Information Processing in Agriculture, 7(2):249–260
Seemanthini K, Manjunath S (2018) Human detection and tracking using hog for action recognition. Procedia Comput Sci 132:1317–1326
Siche R, Vejarano R, Aredo V, Velasquez L, Saldaña E, Quevedo R (2016) Evaluation of food quality and safety with hyperspectral imaging (HSI). Food Eng Rev 8(3):306–322
Silva CS, Sonnadara U (2013) Classification of rice grains using neural networks. Institute of Physics, Sri Lanka, pp 9–14
Szczypiński PM, Klepaczko A, Zapotoczny P (2015) Identifying barley varieties by computer vision. Comput Electron Agric 110:1–8
Taheri-Rad A, Khojastehpour M, Rohani A, Khoramdel S, Nikkhah A (2017) Energy flow modeling and predicting the yield of Iranian paddy cultivars using artificial neural networks. Energy 135:405-412
Timsorn K, Lorjaroenphon Y, Wongchoosuk C (2017) Identification of adulteration in uncooked jasmine rice by a portable low-cost artificial olfactory system. Measurement 108:67–76
Tin MM, Mon KL, Win EP, Hlaing SS (2018) Myanmar rice grain classification using image processing techniques. In: International Conference on big data analysis and deep learning applications. Springer, New York, pp 324–332. https://doi.org/10.1007/978-981-13-0869-7_36
Wang Y, Zhu X, Wu B (2019) Automatic detection of individual oil palm trees from UAV images using hog features and an SVM classifier. Int J Remote Sens 40(19):7356–7370
Ye Q, Yang X, Dai S, Chen G, Li Y, Zhang C (2015) Effects of climate change on suitable rice cropping areas, cropping systems and crop water requirements in southern china. Agric Water Manag 159:35–44
Yin Y, Xiao Y, Yu H (2015) An image selection method for tobacco leave grading based on image information. Eng Agric Environ Food 8(3):148–154
Zareiforoush H, Minaei S, Alizadeh MR, Banakar A (2016) Qualitative classification of milled rice grains using computer vision and metaheuristic techniques. J Food Sci Technol 53(1):118–131
Zayed N, Elnemr HA (2015) Statistical analysis of Haralick texture features to discriminate lung abnormalities. Int J Biomed Imaging 2015
Zhang H, Wang J, Sun Z, Zurada JM, Pal NR (2019) Feature selection for neural networks using group lasso regularization. IEEE Trans Knowl Data Eng:36–49. https://doi.org/0.1109/TKDE.2019.2893266
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Uddin, M., Islam, M.A., Shajalal, M. et al. Paddy seed variety identification using T20-HOG and Haralick textural features. Complex Intell. Syst. 8, 657–671 (2022). https://doi.org/10.1007/s40747-021-00545-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40747-021-00545-0