Abstract
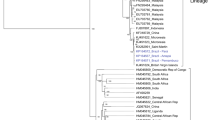
Chikungunya virus (CHIKV) is a mosquito-borne emerging pathogen that is transmitted to humans through the bite of female Aedes mosquitoes. CHIKV infection has become a major public health concern worldwide, as it has a significant impact on the healthcare system. Since 2004, the virus has emerged in Africa and subsequently spread to countries located near the Indian Ocean, including India, and to Europe, the Americas, and Asia. In Thailand, a large CHIKV outbreak occurred during 2008–2009 and was caused by a virus originating from the east/central/south African (ECSA) CHIKV genotype. Since then, the ECSA genotype of CHIKV has continued to circulate and has caused sporadic cases in different areas in Thailand. Approximately 20,000 reported cases have been confirmed by the Bureau of Epidemiology, Ministry of Public Health, Thailand, from January 1, 2018 to July 31, 2020. However, the causes of this CHIKV re-emergence remain unclear. To obtain a better understanding of CHIKV circulation during the recent outbreak in Bangkok, Thailand, complete genome analysis of CHIKV isolates from field-caught mosquitoes collected in outbreak areas was performed. A total of 28 Ae. aegypti samples (21 females and 7 males) were collected, and individual mosquitoes were used for CHIKV detection and isolation. Eleven of 28 (39.29%) female and three of 28 (10.71%) male mosquitoes were positive for CHIKV by E1 nested RT-PCR. Four CHIKV isolates were successfully isolated from four female Ae. aegypti mosquitoes. Based on complete genome analysis, several amino acid substitutions were identified in the protein coding region. The E1:K211E and E2:V264A mutations in the background of the E1:226A mutation were observed in all four CHIKV isolates. An important observation was the presence of one amino acid substitution, leading to an E1:K245R change. This mutation was found in all four CHIKV isolates from mosquitoes in this study and in Thai patients described previously. Additionally, phylogenetic analysis indicated that the four CHIKV isolates belonged to the Indian Ocean clade of the ECSA genotype. The results obtained in this study provide detailed information on the molecular characteristics and evolution of currently circulating CHIKV strains in Thailand, which are useful for developing prevention and control strategies.



Similar content being viewed by others
Data availability statement
The authors confirm that all data underlying the findings are fully available without restriction.
References
Tsetsarkin KA, Chen R, Sherman MB, Weaver SC (2011) Chikungunya virus: evolution and genetic determinants of emergence. Curr Opin Virol 1(4):310–317
Jose J, Snyder JE, Kuhn RJ (2009) A structural and functional perspective of alphavirus replication and assembly. Future Microbiol 4(7):837–856
Chen R, Mukhopadhyay S, Merits A et al (2018) ICTV Virus Taxonomy Profile: Togaviridae. J Gen Virol 99(6):761–762
Ross RW (1956) The Newala epidemic. III. The virus: isolation, pathogenic properties and relationship to the epidemic. J Hyg (Lond) 54(2):177–191
Desdouits M, Kamgang B, Berthet N et al (2015) Genetic characterization of Chikungunya virus in the Central African Republic. Infect Genet Evol 33:25–31
Hammon WM, Rudnick A, Sather GE (1960) Viruses associated with epidemic hemorrhagic fevers of the Philippines and Thailand. Science 131(3407):1102–1103
Rezza G, Nicoletti L, Angelini R et al (2007) Infection with chikungunya virus in Italy: an outbreak in a temperate region. Lancet 370(9602):1840–1846
Halstead SB (2015) Reappearance of chikungunya, formerly called dengue, in the Americas. Emerg Infect Dis 21(4):557–561
Hammon WN, Sather GE (1964) Virological findings in the 1960 hemorrhagic fever epidemic (dengue) in Thailand. Am J Trop Med Hyg 13:629–641
Pialoux G, Gaüzère BA, Jauréguiberry S, Strobel M (2007) Chikungunya, an epidemic arbovirosis. Lancet Infect Dis 7(5):319–327
Ravi V (2006) Re-emergence of chikungunya virus in India. Indian J Med Microbiol 24(2):83–84
Bonilauri P, Bellini R, Calzolari M et al (2008) Chikungunya virus in Aedes albopictus, Italy. Emerg Infect Dis 14(5):852–854
Powers AM, Brault AC, Tesh RB, Weaver SC (2000) Re-emergence of Chikungunya and O’nyong-nyong viruses: evidence for distinct geographical lineages and distant evolutionary relationships. J Gen Virol 81(Pt 2):471–479
Schuffenecker I, Iteman I, Michault A et al (2006) Genome microevolution of chikungunya viruses causing the Indian Ocean outbreak. PLoS Med 3(7):e263
Tsetsarkin KA, Vanlandingham DL, McGee CE, Higgs S (2007) A single mutation in chikungunya virus affects vector specificity and epidemic potential. PLoS Pathog 3(12):e201
Cunha RVD, Trinta KS (2017) Chikungunya virus: clinical aspects and treatment - A Review. Mem Inst Oswaldo Cruz 112(8):523–531
Schwartz O, Albert ML (2010) Biology and pathogenesis of chikungunya virus. Nat Rev Microbiol 8(7):491–500
Tandale BV, Sathe PS, Arankalle VA et al (2009) Systemic involvements and fatalities during Chikungunya epidemic in India, 2006. J Clin Virol 46(2):145–149
Rajapakse S, Rodrigo C, Rajapakse A (2010) Atypical manifestations of chikungunya infection. Trans R Soc Trop Med Hyg 104(2):89–96
Thaikruea L, Charearnsook O, Reanphumkarnkit S et al (1997) Chikungunya in Thailand: a re-emerging disease? Southeast Asian J Trop Med Public Health 28(2):359–364
Pongsiri P, Auksornkitti V, Theamboonlers A et al (2010) Entire genome characterization of Chikungunya virus from the 2008–2009 outbreaks in Thailand. Trop Biomed 27(2):167–176
Tiawsirisup S (2011) Chikungunya virus in Thailand; an update. Thai J Vet Med 41(2):133–134
Rianthavorn P, Prianantathavorn K, Wuttirattanakowit N et al (2010) An outbreak of chikungunya in southern Thailand from 2008 to 2009 caused by African strains with A226V mutation. Int J Infect Dis 3:e161–e165
Theamboonlers A, Rianthavorn P, Praianantathavorn K et al (2009) Clinical and molecular characterization of chikungunya virus in South Thailand. Jpn J Infect Dis 62(4):303–305
Bureau of Epidemiology, Department of Disease Control, MoPH, Thailand (2019) Annual incidence report of Chikungunya virus in Thailand. https://ddc.moph.go.th/dvb/news.php?news=5117&deptcode=dvb. Accessed 3 Aug 2020
Intayot P, Phumee A, Boonserm R et al (2019) Genetic Characterization of Chikungunya Virus in Field-Caught Aedes aegypti Mosquitoes Collected during the Recent Outbreaks in 2019 Thailand. Pathogens 8(3):121
Parida MM, Santhosh SR, Dash PK et al (2007) Rapid and real-time detection of Chikungunya virus by reverse transcription loop-mediated isothermal amplification assay. J Clin Microbiol 45(2):351–357
Hall TA (1999) BIOEDIT: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Trifinopoulos J, Nguyen LT, von Haeseler A, Minh BQ (2016) W-IQ-TREE: a fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res 44(W1):W232–W235
Weaver SC (2014) Arrival of chikungunya virus in the new world: prospects for spread and impact on public health. PLoS Negl Trop Dis 8(6):e2921
Wanlapakorn N, Thongmee T, Linsuwanon P et al (2014) Chikungunya outbreak in Bueng Kan Province, Thailand, 2013. Emerg Infect Dis 20(8):1404–1406
Delatte H, Paupy C, Dehecq JS et al (2008) Aedes albopictus, vector of chikungunya and dengue viruses in Reunion Island: biology and control. Parasite 15(1):3–13
Dzul-Manzanilla F, Martínez NE, Cruz-Nolasco M et al (2016) Evidence of vertical transmission and co-circulation of chikungunya and dengue viruses in field populations of Aedes aegypti (L.) from Guerrero, Mexico. Trans R Soc Trop Med Hyg 110(2):141–144
Heath CJ, Grossi-Soyster EN, Ndenga BA et al (2020) Evidence of transovarial transmission of Chikungunya and Dengue viruses in field-caught mosquitoes in Kenya. PLoS Negl Trop Dis 14(6):e0008362
Thavara U, Tawatsin A, Pengsakul T et al (2009) Outbreak of chikungunya fever in Thailand and virus detection in field population of vector mosquitoes, Aedes aegypti (L.) and Aedes albopictus Skuse (Diptera: Culicidae). Southeast Asian J Trop Med Public Health 40(5):951–962
Jain J, Kushwah RBS, Singh SS et al (2016) Evidence for natural vertical transmission of chikungunya viruses in field populations of Aedes aegypti in Delhi and Haryana states in India-a preliminary report. Acta Trop 162:46–55
Chompoosri J, Thavara U, Tawatsin A et al (2016) Vertical transmission of Indian Ocean Lineage of chikungunya virus in Aedes aegypti and Aedes albopictus mosquitoes. Parasit Vectors 9:227
Phumee A, Intayot P, Sor-Suwan S et al (2021) Molecular detection of Indian Ocean Lineage Chikungunya virus RNA in field collected Culex quinquefasciatus Say from Bangkok, Thailand but no evidence of virus replication. PLoS One 16(1):e0246026
Beerntsen BT, James AA, Christensen BM (2000) Genetics of mosquito vector competence. Microbiol Mol Biol Rev 64(1):115–137
Tsetsarkin KA, Chen R, Yun R et al (2014) Multi-peaked adaptive landscape for chikungunya virus evolution predicts continued fitness optimization in Aedes albopictus mosquitoes. Nat Commun 5:4084
Tsetsarkin KA, McGee CE, Volk SM et al (2009) Epistatic roles of E2 glycoprotein mutations in adaption of chikungunya virus to Aedes albopictus and Ae. aegypti mosquitoes. PLoS One 4(8):e6835
Tsetsarkin KA, Weaver SC (2011) Sequential adaptive mutations enhance efficient vector switching by Chikungunya virus and its epidemic emergence. PLoS Pathog 7(12):e1002412
Tsetsarkin KA, Chen R, Leal G et al (2011) Chikungunya virus emergence is constrained in Asia by lineage-specific adaptive landscapes. Proc Natl Acad Sci U S A 108(19):7872–7877
Agarwal A, Sharma AK, Sukumaran D et al (2016) Two novel epistatic mutations (E1:K211E and E2:V264A) in structural proteins of Chikungunya virus enhance fitness in Aedes aegypti. Virology 497:59–68
Maljkovic Berry I, Eyase F, Pollett S et al (2019) Global outbreaks and origins of a Chikungunya virus variant carrying mutations which may increase fitness for Aedes aegypti: revelations from the 2016 Mandera, Kenya outbreak. Am J Trop Med Hyg 100(5):1249–1257
Patil J, More A, Patil P et al (2018) Genetic characterization of chikungunya viruses isolated during the 2015–2017 outbreaks in different states of India, based on their E1 and E2 genes. Arch Virol 163(11):3135–3140
Shrinet J, Jain S, Sharma A et al (2012) Genetic characterization of Chikungunya virus from New Delhi reveal emergence of a new molecular signature in Indian isolates. Virol J 9:100
Grandadam M, Caro V, Plumet S et al (2011) Chikungunya virus, southeastern France. Emerg Infect Dis 17(5):910–913
Taraphdar D, Chatterjee S (2015) Molecular characterization of chikungunya virus circulating in urban and rural areas of West Bengal, India after its re-emergence in 2006. Trans R Soc Trop Med Hyg 109(3):197–202
Harsha PK, Reddy V, Rao D et al (2020) Continual circulation of ECSA genotype and identification of a novel mutation I317V in the E1 gene of Chikungunya viral strains in southern India during 2015–2016. J Med Virol 92(8):1007–1012
Kaur N, Jain J, Kumar A et al (2017) Chikungunya outbreak in Delhi, India, 2016: report on coinfection status and comorbid conditions in patients. New Microbes New Infect 20:39–42
Agarwal A, Gupta S, Yadav AK et al (2019) Molecular and phylogenetic analysis of Chikungunya virus in Central India during 2016 and 2017 outbreaks reveal high similarity with recent New Delhi and Bangladesh strains. Infect Genet Evol 75:103940
Venturi G, Aberle SW, Avšič-Županc T et al (2020) Specialist laboratory networks as preparedness and response tool - the Emerging Viral Diseases-Expert Laboratory Network and the Chikungunya outbreak, Thailand, 2019. Euro Surveill 25(13):1900438
Feng Y, Xiao H, Li X et al (2020) Nonindigenous East/Central/South African genotype of chikungunya virus identified in febrile returning travellers in Yunnan China. J Infect 80(4):469–496
Phadungsombat J, Imad H, Rahman M et al (2020) A novel sub-lineage of Chikungunya virus East/Central/South African genotype Indian Ocean Lineage caused sequential outbreaks in Bangladesh and Thailand. Viruses 12(11):1319
Fischer C, de Lamballerie X, Drexler JF (2019) Enhanced Molecular Surveillance of Chikungunya Virus. mSphere 4(4):e00295–19
Acknowledgements
We would like to thank the Thailand Research Fund through the Royal Golden Jubilee Ph.D. program (PHD/0054/2560), Ratchadapiseksompote Fund (Grant No. RA63/066), National Research Council of Thailand (NRCT) (Grant No. NRCT5-RSA63001-03), Health Systems Research Institute (Grant No. 64-156), Rachadapisek Sompote Fund (RA/MF 21/63), and NIH/NIAID/CREID/07-049-7012-52338.
Funding
This study was supported by the Thailand Research Fund through the Royal Golden Jubilee Ph.D. program (PHD/0054/2560), Ratchadapiseksompote Fund (Grant No. RA63/066), National Research Council of Thailand (NRCT) (Grant No. NRCT5-RSA63001-03), Health Systems Research Institute (Grant No. 64-156), Rachadapisek Sompote Fund (RA/MF 21/63), and NIH/NIAID/CREID/07-049-7012-52338.
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments: PI, AP, PS. Performed the experiments: PI, AP, SS, RB. Analysed the data: PI, AP, PS. Contributed reagents/materials/analysis tools: PI, AP, KK, PS. Wrote the paper: PI, AP, PS.
Corresponding author
Ethics declarations
Conflict of interest
The authors have declared that they have no competing interests.
Additional information
Handling Editor: Patricia Aguilar.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
705_2021_5243_MOESM1_ESM.png
Supplementary file1 (PNG 140 KB) Supplementary Fig. S1 Alignment of CHIKV E1 amino acid sequences showing the K245R amino acid mutation in the sequences obtained in this study
Rights and permissions
About this article
Cite this article
Intayot, P., Phumee, A., Kraivichian, K. et al. Genetic characterization of chikungunya virus isolates from Aedes aegypti mosquitoes collected during a recent outbreak in Bangkok, Thailand. Arch Virol 166, 3387–3398 (2021). https://doi.org/10.1007/s00705-021-05243-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-021-05243-3