- 1College of Life Science, South-Central University for Nationalities, Wuhan, China
- 2Key Laboratory of Aquatic Botany and Watershed Ecology Wuhan Botanical Garden, Chinese Academy of Sciences, Wuhan, China
- 3Center of the Plant Ecology, Core Botanical Gardens, Chinese Academy of Sciences, Wuhan, China
- 4State Key Laboratory of Agricultural Microbiology, Huazhong Agricultural University, Wuhan, China
Phosphorus (P) availability is a major restriction to crop production, and phosphate-solubilizing bacteria (PSBs) in soils are responsible for P turnover. However, it remains unknown whether the application of PSB can facilitate both inorganic and organic P transformation and enhance function of plant rhizosphere bacteria. In this study, we applied Illumina MiSeq sequencing, plate-colony counting, quantitative PCR, and multiple ecological analyses. We found that the inoculation of PSB Acinetobacter pittii gp-1 significantly promoted the growth of soybean represented by better vegetation properties (e.g., plant height and root P) and increased activities of phosphatase (4.20–9.72 μg/g/h) and phytase (0.69–1.53 μmol/g/day) as well as content of indole acetic acid (5.80–40.35 μg/g/h). Additionally, the application of strain A. pittii gp-1 significantly increased abundances of both inorganic and organic P-cycling-related genes (i.e., phoD, bpp, gcd, and pstS). More importantly, the application of A. pittii gp-1 could increase the function represented by P-cycling-related enzymes (e.g., phosphotransferase) of rhizosphere bacterial community based on functional profiling. To our knowledge, this is the first report that the application of PSB A. pittii promotes inorganic and organic P utilization and increases the function of rhizosphere bacterial community. Therefore, the PSB A. pittii gp-1 could be a good candidate for the promotion of soybean growth.
Introduction
Enhancing the yield of farmland is the most important agricultural issue (Mehrabi and Ramankutty, 2019). P is an essential element for growth and development of plants and, thus, is of significance to the production of fiber and food crops (Hansen et al., 2020; Wan et al., 2020a). At present, the major and wide input of P to farmland is non-renewable P fertilizer, which is often applied beyond the demand of crops due to soil P fixation to metal ions (Neal et al., 2017; Ye et al., 2017). The accumulation of P in soil could lead to the waste of resources and potential environmental risks (e.g., soil compaction and water eutrophication) (Hu et al., 2018). Rational fertilization and improving utilization efficiency of P fertilizer are important agricultural problems.
The transformation of plant-unavailable P (e.g., Ca3(PO4)2, phytate, phospholipid, and nucleic acid) to plant-available P (e.g., H2PO4– and HPO42– ions) needs the participation of P-solubilizing microorganisms (Yu et al., 2011; Wan et al., 2020b). PSBs are responsible for the solubilization of inorganic P and mineralization of organic P (Oliveira et al., 2009; Liu et al., 2014). Phospholipids and phytate are significant organic P pools in soils, which can be hydrolyzed by phosphatase and phytase, respectively (Lim et al., 2007; Maougal et al., 2014; Wei et al., 2019). The inorganic P can be solubilized by small molecular organic acids (e.g., gluconic acid and citric acid), and the formation of small molecular organic acids needs the participation of dehydrogenase (Hanif et al., 2015; Rasul et al., 2019). Previous studies have reported that P-cycling-related genes of phoD, bpp, gcd, and pstS can encode alkaline phosphatase, phytase, glucose dehydrogenase, and phosphatase inorganic transporter system, respectively (Neal et al., 2017; Wan et al., 2020a). Therefore, phoD, bpp, gcd, and pstS genes can be good biomarkers to provide insight into soil P transformation.
Prior studies have reported that specific bacterial community including alkaline phosphomonoesterase-harboring bacterial community and phytase-producing bacterial community can promote plant growth (Maougal et al., 2014; Hanif et al., 2015; Ye et al., 2017; Wei et al., 2019). Additionally, many PSBs have been isolated from natural conditions and found to possess plant growth-promoting capability, such as Acinetobacter (Collavino et al., 2010; Liu et al., 2014), Pseudomonas (Yu et al., 2011), Burkholderia (Collavino et al., 2010), and Bacillus (Hanif et al., 2015; Hansen et al., 2020). The application of PSB in agriculture is a useful approach to enhance soil P availability and avoid excessive use of P fertilizer. Therefore, it is necessary to reveal plant growth-promoting mechanism of PSB. P solubilization and mineralization of single PSB are gradually clarified; however, effects of PSB on transformation of both inorganic and organic P and rhizosphere bacterial community are poorly understood.
To broaden candidates of P-solubilizing microorganisms, we isolated a PSB Acinetobacter pittii gp-1 from agricultural soils (Wan et al., 2020b). In a prior study, we found the strain A. pittii gp-1 showed good performances for utilizing tricalcium phosphate (TCP), aluminum phosphate, iron phosphate, and phytate (Wan et al., 2020b). Soil-derived Acinetobacter bacteria present good P-solubilizing abilities and show great potentials in agroecosystems (Collavino et al., 2010; Yu et al., 2011; Marra et al., 2012; Rasul et al., 2019). However, responses of diversity, composition, and function of indigenous bacterial community to inoculation of PSB Acinetobacter remain unknown. Soybeans are in great demand by human society, and P deficiency leads to poor growth and low production of soybean (Bononi et al., 2020). This situation caught our interest to investigate the growth-promoting capacity of Acinetobacter bacteria for soybean. In the present study, we aimed to (i) investigate effects of PSB inoculation on P transformation and plant growth-promoting performance and (ii) explore responses of soybean rhizosphere bacterial community to the inoculation of PSB. We hypothesized that the inoculation of PSB A. pittii gp-1 would increase P availability and promote the growth of plant and might elevate the P-cycling-related gene abundance. To meet our purpose and address our hypotheses, we conducted potted experiments and Illumina MiSeq sequencing and evaluated soil properties.
Materials and Methods
Potted Experiment Design
The previously isolated PSB A. pittii gp-1 (accession number: MK641660) with indole acetic acid production ability was used in potted experiment. The strain gp-1 was inoculated to 200 ml of the National Botanical Research Institute’s phosphate (NBRIP) medium and incubated at 28°C with shaking of 180 rpm for 5 days. NBRIP medium contained 10 g/L of glucose, 5 g/L of Ca3(PO4)2, 0.25 g/L of MgSO4⋅7H2O, 5 g/L of MgCl2⋅7H2O, 0.2 g/L of KCl, 0.1 g/L of (NH4)2SO4, and 2 ml/L of trace element solution (EDTA, 10 g/L; MnSO4⋅H2O, 2.2 g/L; FeSO4⋅7H2O, 1.0 g/L; CuSO4⋅5H2O, 0.5 g/L; CoCl2⋅6H2O, 0.3 g/L; Na2MoO4⋅2H2O, 0.2 g/L; and CaCl2, 0.1 g/L) (Nautiyal, 1999). After incubation, bacteria were collected by centrifuging and washed three times with sterile water.
The experimental potted soil was collected from an uncultivated field in Wuhan, China (30°28′N, 114°21′E). The soil type is calcareous, with original pH, total carbon, total nitrogen, availability phosphorus, and total phosphorus of 6.9, 0.52, 0.68%, 0.22 mg/g, and 0.89 mg/g, respectively. These P-deficient soils were sieved through a 2-mm mesh to remove stones and plant residuals. TCP was applied as phosphorus source in plant growth promotion experiment as described in previous literatures (Yu et al., 2011; Liu et al., 2014). Four potted treatments were designed: 200 g of sieved soil + 100 ml of sterile water (CK treatment), 195 g of sieved soil + 5 g of TCP + 100 ml of sterile water (Tri treatment), 200 g of sieved soil + 10 ml of bacterial suspension (107 cfu/ml) + 90 ml of sterile water (Sup treatment), and 195 g of sieved soil + 5 g of TCP + 10 ml of bacterial suspension (107 cfu/ml) + 90 ml of sterile water (Bac treatment). Each treatment had five replications. Soybean seeds (Glycine max w82) were purchased from China National Seed Group, pre-cultivated in sterile nutritious soils, and allowed them grow to about 10-cm length of sprouts. Each sprout with same growth potential was transplanted to each plastic pot as described above, and the strain gp-1 was inoculated to soybean rhizosphere in Sup and Bac treatments. Each plot was covered with Nylon membrane. These pots were randomly placed in greenhouse and incubated at 25°C with the cycling treatment of 16-h light and 8-h dark for a total of 40 days.
Determination of Phosphate-Solubilizing Bacterium Abundance and Indole Acetic Acid
Every 10 days, we used alcohol-wiped shovels and tweezers to collected about 5 g of bulk soils near soybean root from each pot. In the experiment of plate-colony counting for abundance of PSB, 1 g of freeze-dried soil was added to 10 ml of sterile water and shaken at 180 rpm for 30 min, and the mixture is allowed to stand for 10 min. Then 1 ml of soil suspension was diluted, 0.1 ml × 10–6 of diluent was evenly spread on NBRIP solid medium containing 0.2 g/L of cycloheximide acting as fungicide and incubated at 28°C for 5 days. After incubation, the cfu in different plates were counted. We also estimated content of indole acetic acid by using Van Urk Salkowski reagent, and the standard approach has been described previously (Biswas et al., 2018).
Determination of Soil Physicochemical Properties, Enzyme Activity, and Vegetation Properties
After 40-day growth of soybean, we excluded pots with the best and worst soybean growth in each treatment, and then 12 pots were left. We scraped rhizosphere soils by using a brush. We measured soil physicochemical properties, including pH, total carbon, total nitrogen, and available P, based on standard methods (Wan et al., 2021a). Microbial biomass P was evaluated by chloroform fumigation extraction and was calculated as the difference between fumigated and non-fumigated subsamples and simultaneously revised for the incomplete recovery of a P spike (Roberts et al., 2013; Ragot et al., 2016).
Soil alkaline phosphatase activity and phytase activity were determined according to previous methods (Wan et al., 2020a). Phosphatase activity and phytase activity were expressed as μg pNPP produced per gram of freeze-dried soil in 1 h and μmol P produced per gram of freeze-dried soil in 1 day, respectively.
The pots in each group was kept to measure the plant height, plant fresh weight, plant dry weight, leaf number, leaf fresh weight, root length, and root fresh weight. Soybean shoots and roots were separated from plants and dried at 60°C. The clean and dried root and shoot were separately cut into small pieces and digested by concentrated H2SO4–H2O2. The digested solutions were applied for measuring the content of root P and shoot P (Fraser et al., 2017).
DNA Extraction, Gene Quantification, Amplicon Sequencing, and Sequence Processing
Three rhizosphere soils from each group were used to extract total DNA using a DNA extraction kit (Mo Bio, Carlsbad, CA, United States) according to the manufacturer’s instruction. DNA concentrations were determined using a NanoDrop 2,000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, United States). All extracted DNA samples were stored at –80°C.
The absolute abundances of phosphorus-cycling-related genes in soil bacteria were measured using qPCR with SYBR green mix. Primer sequences for amplifying P-cycling-related genes (i.e., phoD, bpp, gcd, and pstS) and quantitation PCR condition are summarized in Supplementary Method 1. Additionally, we used these primers to amplify bpp, phoD, gcd, and pstS from A. pittii gp-1.
The V3–V4 region of bacterial 16S rRNA gene was amplified using the primers 338F (5′-ACT CCT ACG GGA GGC AGC A-3′) and 806R (5′-GGA CTA CHV GGG TWT CTA AT-3′) (Mori et al., 2013). A PCR of 20 μl was performed in triplicate using a thermal cycler (ABI 9700, Thermo, United States) and conducted at the following conditions: an initial denaturation at 95°C for 3 min, 30 cycles of 95°C for 40 s, 58°C for 40 s, and 72°C for 50 s, and then a final extension at 72°C for 10 min. Sequencing was conducted on an Illumina MiSeq platform at Majorbio Bio-Pharm Technology Co., Ltd., Shanghai, China.
The raw reads were processed to gain purified sequences following the pathway of QIIME (Caporaso et al., 2010). We eliminated (1) sequences that did not exactly match barcodes and primers; (2) sequences with an average quality score < 20; (3) sequences with maximum homopolymers < 10 bp; and (4) sequences that contained ambiguous bases call. The purified sequences were clustered into operational taxonomic units (OTUs) at 97% identity against the SILVA v128 reference set.
Statistical Analysis
Significant differences were calculated by the one-way analysis of variance with means compared using Tukey’s test in R. Venn diagram and non-metric multidimensional scaling (NMDS) plot were used to reflect bacterial community composition. Pairwise analysis of similarity (ANOSIM) was applied to quantitatively evaluate difference in bacterial community composition by using the ‘‘anosim’’ function in the ‘‘vegan’’ package of R. Permutational multivariate analysis of variance (PERMANOVA) was applied to evaluate pure effect of factors (e.g., physicochemical parameters and enzyme activity) on vegetation properties by using the ‘‘adonis’’ function in the ‘‘vegan’’ package of R. Linear discriminant analysis (LDA) effect size (LEfSe) statistical analysis was conducted on the online interface Galaxy1 at a significant level of p < 0.05 and an LDA score > 4. Functional profiling of bacterial taxa was carried out by applying the “Tax4Fun2” package in R, and the functional redundancy index for each sample was calculated based on 16S rRNA gene similarity (Wemheuer et al., 2020). Canonical analysis of principal coordinates was applied to investigate influences of components including soil physicochemical parameters, gene abundance, cell exudates (include enzyme and indole acetic acid), and relative abundances of phylum bacteria on the vegetation properties. To identify core taxa, OTUs observed in more than 50% of all samples (> 6 samples, 875 OTUs) were applied to build a co-occurrence network. The co-occurrence network was visualized using Gephi v. 0.9.22 at a significant level of p < 0.01 and Spearman’s correlation coefficient higher than 0.67 (Wan et al., 2021b). Structural equation model was built to show relationships among vegetation properties, physicochemical properties, gene abundance, cell exudate, and bacterial community composition by using the packages of “sem” and “plspm” in R. The first principal component (PC1) value of soil physicochemical properties, P-cycling-related gene abundance, bacterial community composition, cell exudate, and vegetation properties accounting for 96.19, 85.19, 41.56, 98.99, and 96.37% of the total variances, respectively, were used as a proxy in structural equation model.
Results
Shifts in Phosphate-Solubilizing Bacterium Abundance and Indole Acetic Acid Content During Soybean Growth
The PSB abundance represented by the number of cfu showed significant difference in four treatments (CK, Tri, Sup, and Bac) during 40-day growth of soybean (Figure 1A). The abundance of PSB in Bac treatment dramatically increased from 3.57 × 107 cfu/g soil at day 10 to 6.96 × 107 cfu/g soil at day 40 (p < 0.05). The population of PSB fluctuated in CK, Tri, and Sup treatments during 40 days but did not significantly ascend at day 40 than at day 10 (p > 0.05). The abundance of PSB in Bac treatment was significantly higher than that in other groups; this difference might be partially due to the input of A. pittii gp-1 and TCP. In addition, we randomly picked 10 colonies from the plate and found that 16S rRNA gene sequence of three bacterial colonies presented 100% similarity with that of A. pittii gp-1.
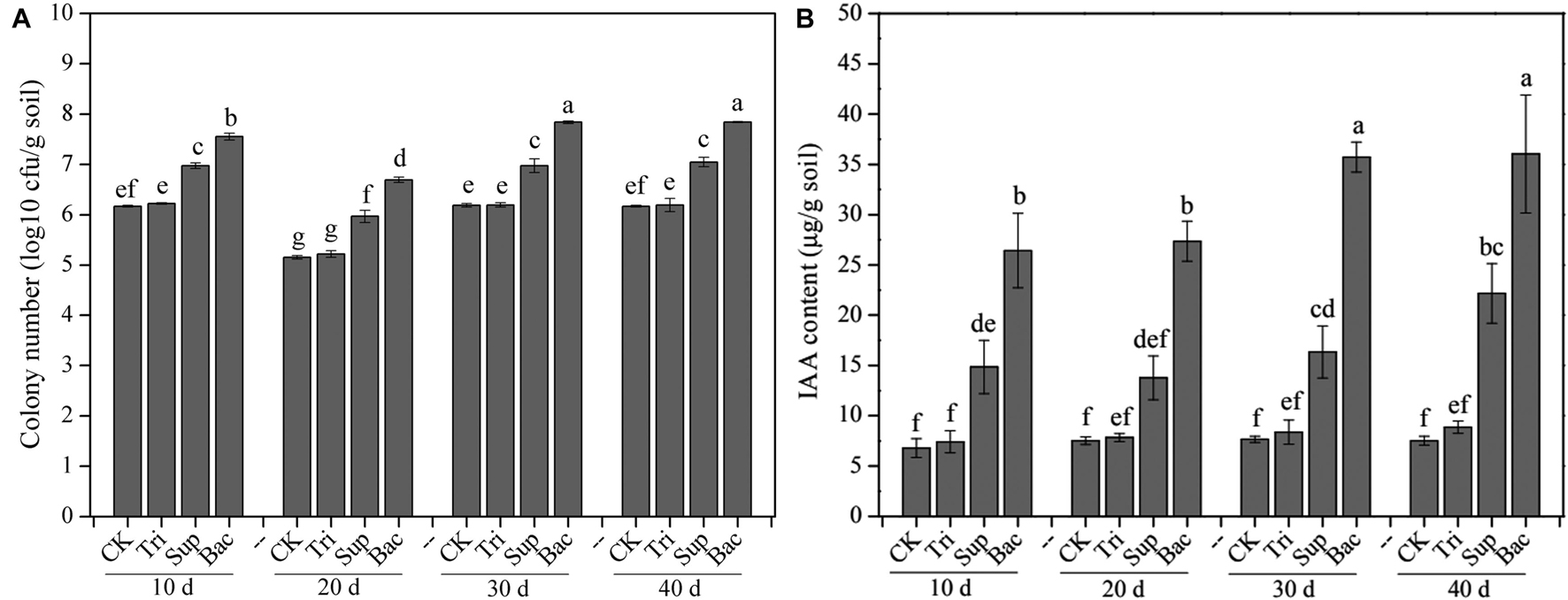
Figure 1. The colony-forming unit number of phosphate-solubilizing bacteria (A) and content of indole acetic acid (B) during 40 days. The results are the mean value of five replicates; error bars represent standard error. Different letters above the column indicate significance (p < 0.05).
The indole acetic acid content was significantly higher in Bac treatment than in other treatments in each period (p < 0.05; Figure 1B). Additionally, the indole acetic acid content noticeably increased in Bac and Sup treatments during 40 days (p < 0.05), while in CK and Tri treatments, it did not (p > 0.05). Linear regression indicated that abundance of PSB was significantly correlated with content of indole acetic acid (Supplementary Figure 1). This suggests that PSB could produce and release indole acetic acid, which in turn might promote soybean growth.
Vegetation Properties, Soil Physicochemical Properties, and P-Cycling-Related Gene Abundance
After 40 days’ growth, the soybean presented erect leaves that became dark green (Supplementary Figure 2). Differences in vegetation properties were found in four treatments, including the plant height, plant fresh weight, plant dry weight, leaf number, leaf fresh weight, root length, root fresh weight, shoot P, and root P (Table 1). Plant dry weight, root length, shoot P, and root P were significantly higher in Bac group than in other three groups (p < 0.05). More importantly, the plant length, plant fresh weight, plant dry weight, leaf number, leaf fresh weight, root length, root fresh weight, root P, and shoot P were dramatically higher in Sup treatment than in CK treatment (p < 0.05). This suggests that the inoculation of PSB A. pittii gp-1 promotes soybean growth.
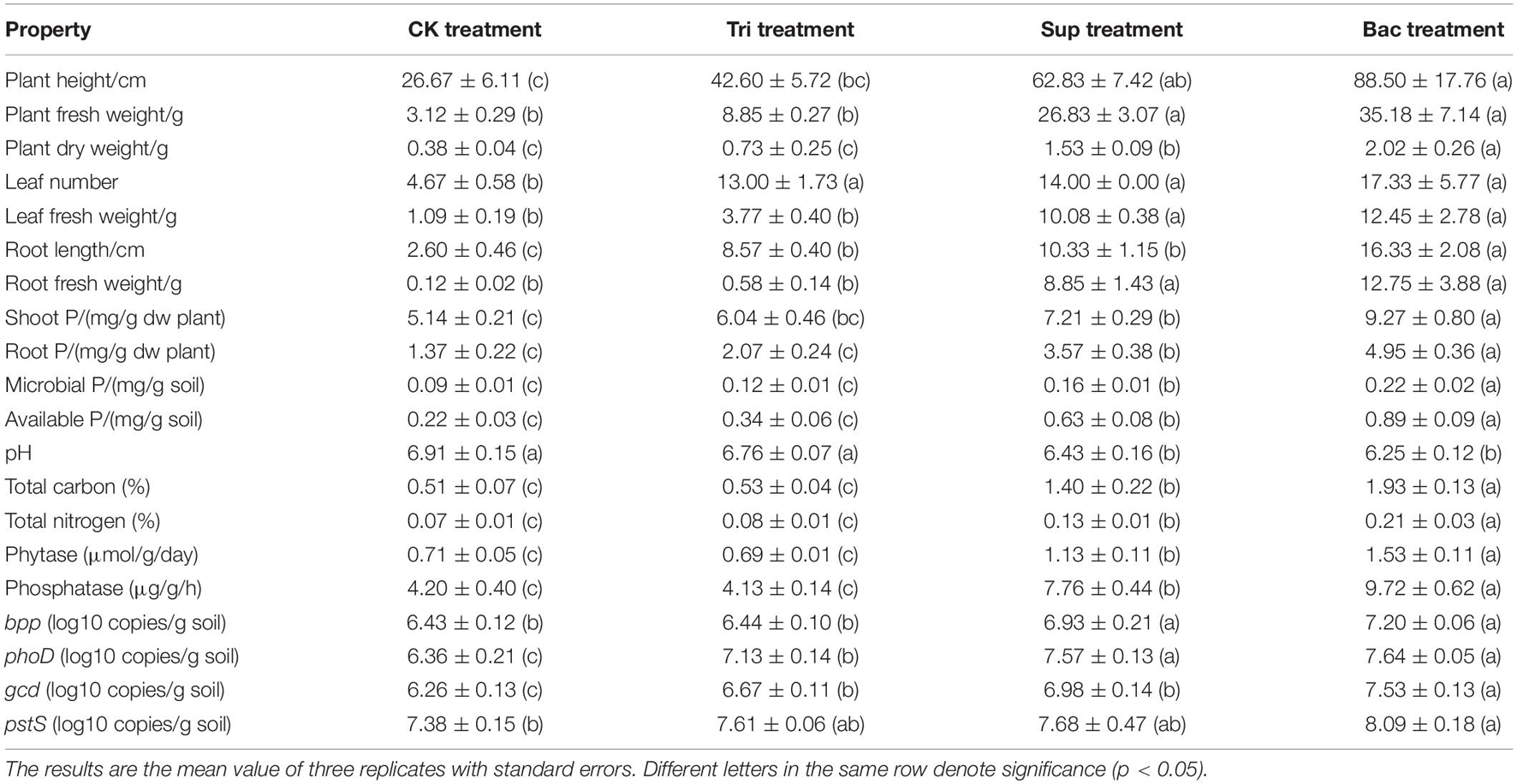
Table 1. Vegetation properties, soil physicochemical properties, enzyme activity, and P-cycling-related gene abundance in four potted treatments.
The soil pH (6.1–7.0) was significantly lower in Bac and Sup treatments than that in CK and Tri treatments (Table 1). Total carbon (0.43–2.04%), total nitrogen (0.07–0.24%), available P (0.19–0.99 mg/g), microbial biomass P (0.08–0.24 mg/g), alkaline phosphatase activity (3.79–10.25 μg/g/h), and phytase activity (0.66–1.63 μmol/g/day) were remarkably higher in Bac treatment than in other treatments (p < 0.05). These results indicate that the inoculation of PSB A. pittii gp-1 increases P availability and microbial activity.
Basically, the abundances of bpp, phoD, gcd, and psts genes were higher in Bac and Sup treatments than in CK and Tri treatments (Figure 1B). Expectedly, the abundance of gcd was significantly higher in Bac treatment (3.36 × 107 copies/g soil) than that in CK treatment (1.86 × 106 copies/g soil), Tri treatment (4.68 × 106 copies/g soil), and Sup treatment (9.62 × 106 copies/g soil). Linear regressions reflected significantly positive correlations between phoD gene abundance and alkaline phosphatase activity (R2 = 0.585, p < 0.01), between bpp gene abundance and phytase activity (R2 = 0.892, p < 0.001), and between gcd gene abundance and indole acetic acid content (R2 = 0.854, p < 0.001) (Supplementary Figure 3). Additionally, gcd and pstS could be amplified from strain A. pittii gp-1 using primers described above, while bpp and phoD did not. These results might imply that the addition of A. pittii gp-1 could increase the abundances of organic P-cycling-related bacterial abundance.
General Properties of Rhizosphere Bacterial Community
A total of 2,829 OTUs were found across 12 soil samples. The CK, Tri, Sup, and Bac treatments possessed 1,670, 1,556, 1,413, and 906 OTUs, respectively; and they shared 181 OTUs (Figure 2A). A total of 39 phyla were observed, and 11 phyla with relative abundance > 0.01% were found across these 12 samples (Figure 2B). Proteobacteria, Chloroflexi, Actinobacteria, and Firmicutes were the first level dominant bacteria, with corresponding relative abundance from 7.94 to 48.22%, from 0.92 to 46.58%, from 8.86 to 44.47%, and from 1.13 to 25.25%, respectively. Acidobacteria, Bacteroidetes, Cyanobacteria, Deinococcus–Thermus, Gemmatimonadetes, Nitrospirae, and Saccharibacteria were the secondary dominant bacteria. The NMDS result showed that distinct difference in bacterial community composition among four treatments (Figure 2C). ANOSIM confirmed further the significant difference (R = 0.6451, p < 0.001). According to LEfSe result, bacteria including Bacillus and Acinetobacter were dramatically abundant in Bac treatment, while bacteria including Acinetobacter, Nitrospira, and Rhodobacter were significantly abundant in Sup treatment (Supplementary Figure 4). According to PERMANOVA results, the application of TCP explained 23.40% of the total variation in community composition (F = 5.27, p < 0.01), and the application of A. pittii gp-1 explained 29.11% of the total variation in community composition (F = 5.75, p < 0.001).
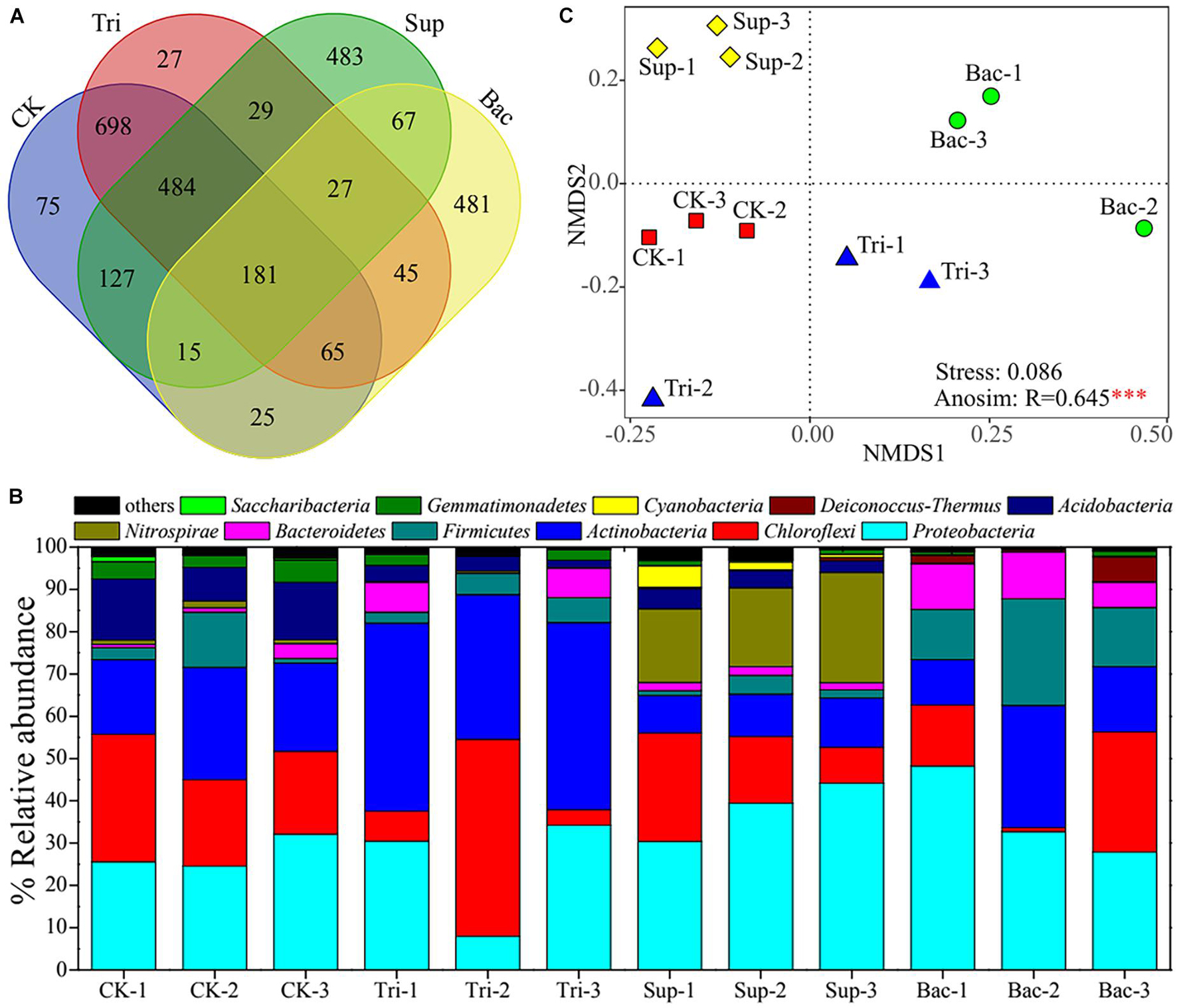
Figure 2. Composition of rhizosphere bacterial community. (A) Venn diagram shows the shared core microbiomes among four groups. (B) Stacking diagram reflects relative abundances of top 11 bacterial phyla (relative abundance > 1%) in 12 soil samples. (C) Non-metric multidimensional scaling plot exhibits difference in bacterial community composition among four treatments. Asterisks denote significance (∗∗∗p < 0.001).
The bacterial community diversity represented by the Shannon–Wiener index (3.85–6.16) and community richness represented by Chao1 index (687–1303) were significantly lower in Bac treatment than in other treatments (p < 0.05; Supplementary Figure 5). This suggests that the addition of A. pittii gp-1 and TCP decreased rhizosphere bacterial diversity.
Based on functional profiling results, 3,113 functions at Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway level 3, including carbon-, nitrogen-, phosphorus-, and sulfate-cycling-related enzymes or proteins, displayed a higher functional redundancy in CK + Tri (without A. pittii gp-1 addition), whereas 3,772 functions had higher redundancies in Sup + Bac (with A. pittii gp-1 addition) (Figure 3A). It was worth noting that 206 functions representing P-cycling-related enzymes or proteins were higher in Sup + Bac than in CK + Tri, such as phosphoglycerate dehydrogenase (EC: 1.1.1.95) and phosphoglycerate kinase (EC: 2.7.2.3). Additionally, 35 functions [(e.g., L-iduronidase (EC: 3.2.1.76), dCTP deaminase (EC: 3.5.4.30), and phloroglucinol synthase (EC: 2.3.1.253)] were unique in CK + Tri, while 198 functions [e.g., phosphotransferase (EC: 2.7.1.-), neamine phosphoribosyltransferase (EC: 2.4.2.49), 5-phosphoribostamycin phosphatase (EC: 3.1.3.88), and uracil phosphatase (EC: 3.1.3.104)] were exclusive in Sup + Bac. At KEGG level 2, some functions (e.g., metabolism of cofactors and vitamins, energy metabolism, and translation) were significantly higher in Sup + Bac than in CK + Tri (p < 0.05), but some functions were not (Figure 3B).
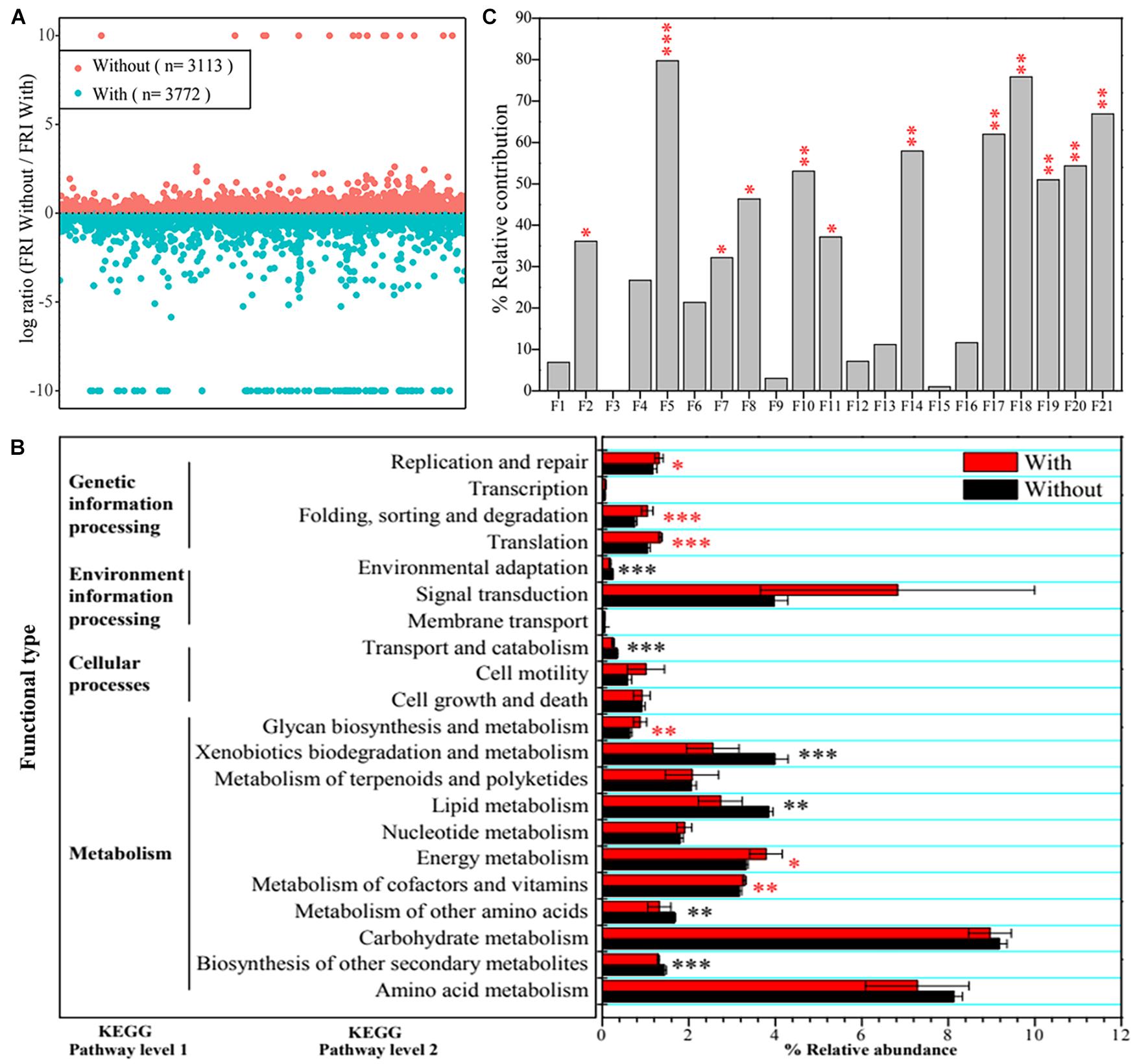
Figure 3. Community functional differences and functional contributions to vegetation property. (A) Functional redundancy indices (FRIs) of bacterial community in soils with inoculation of phosphate-solubilizing bacteria (PSBs) (Sup + Bac) and soils without addition of PSBs (CK + Tri) soils. A log ratio > 0 denotes that a function is more redundant in soils without PSB addition. (B) Differences in bacterial functions between group with addition of PSBs (Sup + Bac) and group without addition of phosphorus-solubilizing bacteria (CK + Tri) at Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway level 1 and level 2. (C) Effects of functions at KEGG pathway level 2 on vegetation property determined by permutational multivariate analysis of variance (PERMANOVA). The abbreviations of F1–F21 represent functions in panel B (from bottom to up, namely, from amino acid metabolism to replication and repair). Asterisks denote significance (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001).
A co-occurrence network was constructed to reveal the relationships among bacterial taxa (Figure 4A). We found 50,510 positive edges (represent significantly positive correlation) and two negative edges (denote dramatically negative correlation), suggesting that rhizosphere bacteria presented a less conflicting interaction. We also clarified the top 20 core nodes; i.e., those with the highest betweenness centrality were affiliated with Acidobacteria (e.g., OTU522), Actinobacteria (e.g., OTU947), Chloroflexi (OTU67), Firmicutes (e.g., OTU601), Gemmatimonadetes (OTU1967), and Proteobacteria (e.g., OTU1813) (Figure 4B).
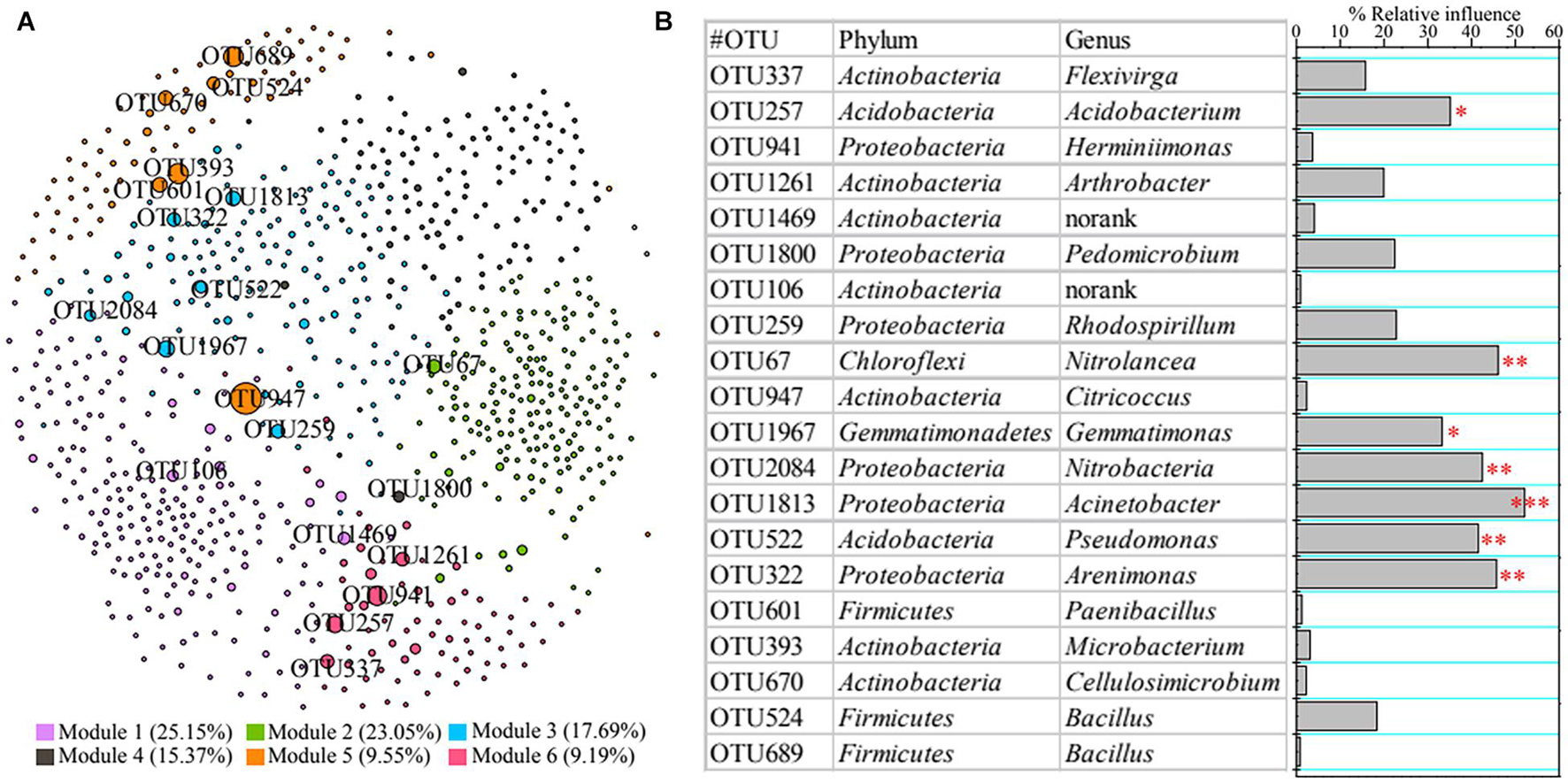
Figure 4. Co-occurrence network of rhizosphere bacteria (A) and contributions of core taxa to vegetation property based on permutational multivariate analysis of variance (PERMANOVA) (B). Asterisks denote significance (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001).
Effects of Abiotic and Biotic Factors on Vegetation Properties
According to PERMANOVA results, the application of TCP could explain 13.69% of the total variation (F = 20.23, p < 0.01) in vegetation properties, while the application of A. pittii gp-1 could explain 72.41% of the total variation (F = 107.05, p < 0.001). According to results of canonical analysis of principal coordinates, soil physicochemical properties (Figure 5A), gene abundance (Figure 5B), cell exudates (Figure 5C), and relative abundances of bacterial phyla (Figure 5D) explained more than 80% of the total variation in vegetation properties. Physicochemical parameter, gene abundance, enzyme activity and IAA, and relative abundance of bacterial phylum showed significantly pure effects on vegetation properties based on PERMANOVA (Figure 5).
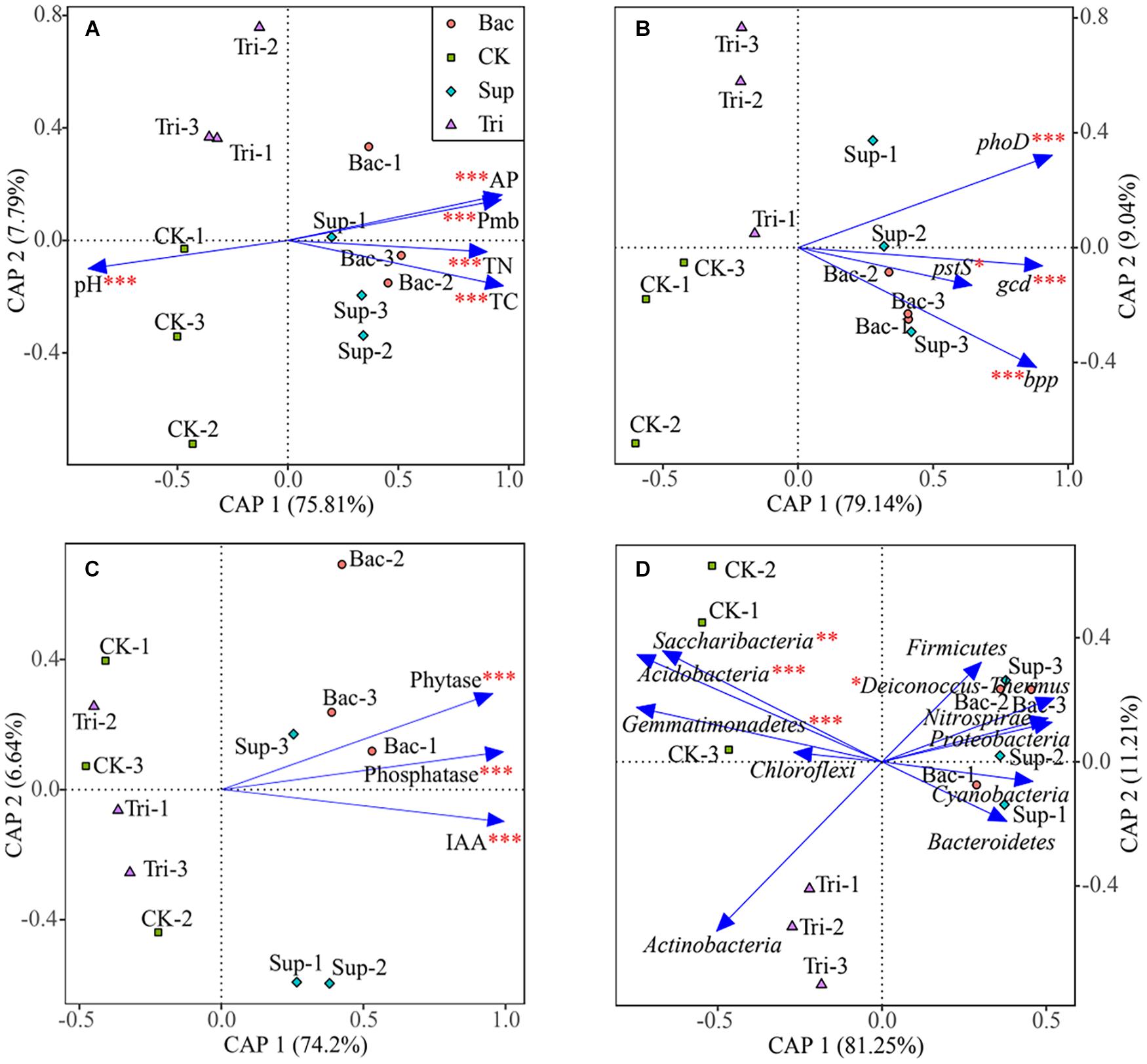
Figure 5. Canonical analysis of principal coordinates showing effects of soil physicochemical properties (A), abundances of phosphorus-cycling-related genes (B), cell exudates (C), and bacterial abundances at phylum level (D) on vegetation properties. The significance of factors was determined using permutational multivariate analysis of variance (PERMANOVA) and is reflected by asterisks next to the variable names. Asterisks denote significance (∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001).
Additionally, we also found that bacterial functions based on functional profiling were responsible for vegetation properties (Figure 3C). The function of metabolism of cofactors and vitamins (R2 = 79.75%, F = 39.38; p < 0.01) showed greater effect on vegetation properties than other functions according to PERMANOVA results. The core taxa identified from co-occurrence network also have significant effects on vegetation properties based on PERMANOVA (Figure 4B). The OTU1813 regarded as Acinetobacter genus presented higher influence (R2 = 52.08%, F = 10.87; p < 0.01) than other core taxa.
Ultimately, we used structural equation model to reveal interconnections among soil physicochemical properties, P-cycling-related gene abundance, bacterial community composition, enzyme activity, and vegetation properties (Figure 6). The model presented a good fit to our data, as indicated by the non-significant χ2-test (N = 12, χ2 = 0.707, d.f. = 1, p = 0.400). On the one hand, bacterial community could affect soil physicochemical properties and P-cycling-related gene abundance, which in turn affect vegetation properties; on the other hand, soil physicochemical properties and P-cycling-related gene abundance could influence enzyme activity, which in turn influences vegetation properties. These results indicated that soil, plant, and bacteria presented close relationships.
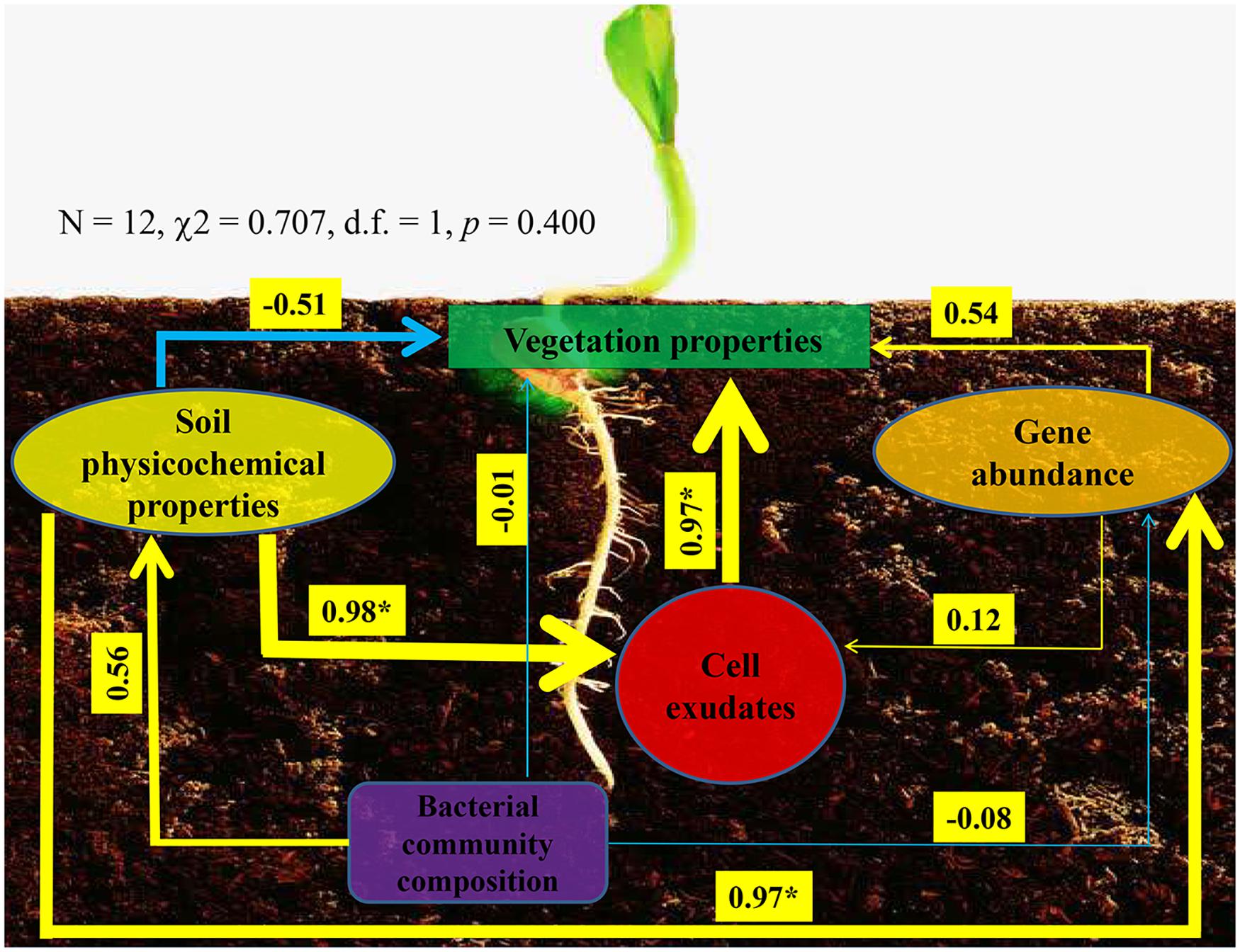
Figure 6. Structural equation model showing the hypothesized causal relationships among vegetation properties, soil physicochemical properties, phosphorus-cycling-related gene abundance, cell exudates (include enzyme and indole acetic acid), and bacterial community composition. The width of the arrows presents the strength of the standardized path coefficient. The blue lines indicate negative path coefficients, and yellow lines reflect positive path coefficients. Values above the lines indicate path coefficients between two parameters. Asterisks denote significance (p < 0.05).
Discussion
Promoting efficient utilization of P is important in agriculture due to rapidly increasing cost of fertilizers and big concerns of environmental protection (Hu et al., 2018). The bacteria possessing P utilization capacity are widespread in the rhizosphere soils of different crops (Maougal et al., 2014; Hanif et al., 2015; Wan et al., 2020b) and promise great application potentials in agriculture because PSB are responsible for P availability and facilitate P uptake by crops (Richardson et al., 2011; Bononi et al., 2020; Pastore et al., 2020). However, the activity and abundance of PSB are subjected to the fertilization treatment and phosphorus fractions (Luo et al., 2017; Hu et al., 2018; Wei et al., 2019; Wan et al., 2020a). Therefore, the isolation and application of highly efficient PSB are meaningful in terms of promoting soil P availability in agroecosystems.
Elucidating Soybean Growth-Promoting by Phosphate-Solubilizing Bacteria Acinetobacter Pittii gp-1
Applying PSB can increase soil available P content (Maougal et al., 2014) and promote vegetation growth (Yu et al., 2011; Biswas et al., 2018; Hansen et al., 2020). However, it should be considered whether the PSB could maintain their activity, function, and abundance after inoculation. In this study, the inoculation of PSB A. pittii gp-1 significantly promoted the growth of soybean represented by better vegetation properties, which is in accordance with prior findings describing that PSB can enhance the growth of legume plant (Collavino et al., 2010; Bononi et al., 2020; Cumpa-Velásquez et al., 2021) and other kinds of plants (Yu et al., 2011; Liu et al., 2014). In these studies, the increase in the content of available P or small molecular organic acid is closely correlated with the growth of plants. The PSB Acinetobacter genus is reported to have the ability to release small molecular organic acid (e.g., indole acetic acid, gluconic acid, oxalic acid, and citric acid) (Marra et al., 2012; Marwa et al., 2019; Rasul et al., 2019). Interestingly, we found the A. pittii gp-1 could produce indole acetic acid detected by using the Van Urk Salkowski reagent. Therefore, the inoculation of the A. pittii gp-1 might increase the content of soil organic acid, which in turn increased the content of available P. Additionally, we detected Acinetobacter genus in Bac treatment by using simple 16S rRNA gene sequencing for single colony. Illumina MiSeq sequencing result also reflected that Acinetobacter dominated in Sup and Bac groups. These results suggest that A. pittii gp-1 could survive after inoculation and could promote the growth of soybean.
In addition, we used four pairs of primers as described above to amplify bpp, phoD, gcd, and pstS genes from A. pittii gp-1. Unexpectedly, only gcd and pstS genes could be amplified. Previous studies have reported that Acinetobacter genus harbors gcd and pstS gene (Marra et al., 2012; Farrugia et al., 2015; Wan et al., 2020b), and almost no study has reported that Acinetobacter genus possesses bpp and phoD genes. However, the abundances of P-cycling-related genes including bpp, phoD, gcd, and pstS were higher in Sup and Bac treatments. These results and findings suggest that the inoculation of PSB A. pittii gp-1 might significantly increase both inorganic and organic P-cycling-related gene abundance of soil indigenous bacteria. This phenomenon might be due to the solubilization of inorganic P by added PSB A. pittii gp-1 via releasing small molecular organic acid. Consequently, part of soluble P was assimilated by native bpp-harboring bacteria and phoD-harboring bacteria and in turn enriched the abundances of bpp and phoD genes and released more phosphatase and phytase. In addition, a part of inoculated A. pittii gp-1 might die; thus, the cell residues could be treated as nutrient for indigenous microorganisms. Previous literatures have reported that gcd-harboring bacteria can produce and release small organic acid to solubilize insoluble inorganic P, thus promoting the growth of plant (Wagh et al., 2014; Rasul et al., 2019). The bpp-harboring bacteria and phoD-harboring bacteria are reported to be responsible for the turnover of soil organic P by releasing extracellular enzyme, which in return promotes the growth of vegetation (Maougal et al., 2014; Ragot et al., 2016; Hu et al., 2018; Zhang et al., 2021). Therefore, the application of PSB A. pittii gp-1 could enhance utilization potentials of both inorganic and organic P.
Response of Rhizosphere Bacterial Community to Inoculation of Strain gp-1
Considering community diversity is closely correlated with soil ecosystem functions (Wan et al., 2021c), it is important to decipher effects of the application of PSB on plant rhizosphere bacterial community. We found significant decrease in rhizosphere bacterial diversity and distinct change in bacterial community composition, which is similar to findings in published literatures (Estrada-Bonilla et al., 2017; Wei et al., 2017; Widdig et al., 2019). In addition, earlier studies have reported that vegetation also affects the composition of bacterial community (Xue et al., 2017; Yang et al., 2018; Campos-Herrera et al., 2019). To the best of our knowledge, this is the first report that the addition of PSB A. pittii could promote the community function of rhizosphere bacteria especially phosphorus-cycling-related functions. This phenomenon might be due to elevated nutrient caused by inoculation of PSB A. pittii, which in turn affected rhizosphere bacterial community composition and function. An earlier study has reported that dead bacteria can be treated as available nutrient to affect growth of other microorganisms (Hanajima et al., 2019). Additionally, microbial biomass P contributes to P solubility in riparian vegetated buffer strip soils (Roberts et al., 2013).
Based on these results and findings, we raised one question of whether there were close relationships among plant, soil, and rhizosphere. The structural equation model reflected stronger interconnections among vegetation properties, soil physicochemical properties, P-cycling-related gene abundance, cell exudates, and bacterial community composition. This result is similar to our prior finding (Wan et al., 2021a). The co-occurrence network also showed that core taxa belonging to Acidobacteria, Chloroflexi, Gemmatimonadetes, and Proteobacteria presented significant effects on vegetation properties. Previous literature has reported that some specific phylum bacteria, such as Acidobacteria, Actinobacteria, and Proteobacteria, are responsible for vegetation growth under different P conditions (Bergkemper et al., 2016). Vegetation properties and microbes could also affect each other (Neal et al., 2017; Yang et al., 2018; Muñoz et al., 2021), suggesting that soil, plant, and bacteria have close relationships. In the future, we will explore molecular mechanisms to reveal close interconnections among soil, plant, and bacteria.
Conclusion
The application of TCP and A. pittii gp-1 could significantly increase soil available P, enrich both inorganic and organic P-cycling-related gene abundance, and promote the growth of soybean. Addition of TCP and A. pittii gp-1 significantly alters the local bacterial community composition after 40-day soybean growth. To our knowledge, we firstly report that the addition of Acinetobacter could promote both inorganic and organic P utilization and could increase the function of rhizosphere bacterial community. Phosphate-solubilizing bacterium A. pittii gp-1 could be a good candidate for the growth promotion of soybean in agroecosystems, and experiments will be conducted to estimate its growth-promoting performance for more different plants in future studies.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, SRR8742689–SRR8742700.
Author Contributions
WW and DH designed the whole experiments. WW conducted all the experiments, analyzed the data, and wrote the manuscript. DH revised the manuscript. Both authors contributed to the article and approved the submitted version.
Funding
This work was supported by grants from the National Natural Science Foundation of China (Grant No. 31772399) and the Fundamental Research Funds for the Central Universities (Grant No. 2662015PY116).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.737116/full#supplementary-material
Footnotes
References
Bergkemper, F., Schöler, A., Engel, M., Lang, F., Krüger, J., Schloter, M., et al. (2016). Phosphorus depletion in forest soils shapes bacterial communities towards phosphorus recycling systems. Environ. Microbiol. 18, 1988–2000. doi: 10.1111/1462-2920.13188
Biswas, J. K., Banerjee, A., Rai, M., Naidu, R., Biswas, B., Vithanage, M., et al. (2018). Potential application of selected metal resistant phosphate solubilizing bacteria isolated from the gut of earthworm (Metaphire posthuma) in plant growth promotion. Geoderma 330, 117–124. doi: 10.1016/j.geoderma.2018.05.034
Bononi, L., Chiaramonte, J. B., Pansa, C. C., Moitinho, M. A., and Melo, I. S. (2020). Phosphorus-solubilizing Trichoderma spp. from Amazon soils improve soybean plant growth. Sci. Rep. 10:2858. doi: 10.1038/s41598-020-59793-8
Campos-Herrera, R., Blanco-Perez, R., Bueno-Pallero, F. A., Duarte, A., Nolasco, G., Sommer, R. J., et al. (2019). Vegetation drives assemblages of entomopathogenic nematodes and other soil organisms: evidence from the Algarve, Portugal. Soil Biol. Biochem. 128, 150–163. doi: 10.1016/j.soilbio.2018.10.019
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of highthroughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Collavino, M. M., Sansberro, P. A., Mroginski, L. A., and Aguilar, O. M. (2010). Comparison of in vitro solubilization activity of diverse phosphatase-solubilizing bacteria native to acid soil and their ability to promote Phaseolus vulgaris growth. Biol. Fert. Soils 46, 727–738. doi: 10.1007/s00374-010-0480-x
Cumpa-Velásquez, L. M., Moriconi, J. I., Dip, D. P., Castagno, L. N., Puig, M. L., Maiale, S. J., et al. (2021). Prospecting phosphate solubilizing bacteria in alkaline-sodic environments reveals intra-specific variability in Pantoea eucalypti affecting nutrient acquisition and rhizobial nodulation in Lotus tenuis. Appl. Soil. Ecol. 168:104125. doi: 10.1016/j.apsoil.2021.104125
Estrada-Bonilla, G. A., Lopes, C. M., Durrer, A., Alves, P. R. L., Passaglia, N., and Cardoso, E. J. B. N. (2017). Effect of phosphate-solubilizing bacteria on phosphorus dynamics and the bacterial community during composting of sugarcane industry waste. Syst. Appl. Microbiol. 40, 308–313. doi: 10.1016/j.syapm.2017.05.003
Farrugia, D. N., Elbourne, L. D. H., Mabbutt, B. C., and Paulsen, L. T. (2015). A novel family of integrases associated with prophages and genomic islands integrated within the tRNA-dihydrouridine synthase A (dusA) gene. Nucleic Acids Res. 43, 4547–4557. doi: 10.1093/nar/gkv337
Fraser, T. D., Lynch, D. H., Gaiero, J., Khosla, K., and Dunfield, K. E. (2017). Quantification of bacterial non-specific acid (phoC) and alkaline (phoD) phosphatases genes in bulk and rhizosphere soil from organically managed soybean fields. Appl. Soil Ecol. 111, 48–56. doi: 10.1016/j.apsoil.2016.11.013
Hanajima, D., Aoyagi, T., and Hori, T. (2019). Dead bacterial biomass-assimilating bacterial populations in compost revealed by high-sensitivity stable isotope probing. Environ. Int. 133:105235. doi: 10.1016/j.envint.2019.105235
Hanif, M. K., Hameed, S., Imran, A., Naqqash, T., Shahid, M., and Van Elsas, J. D. (2015). Isolation and characterization of a β-propeller gene containing phosphobacterium Bacillus subtilis strain KPS-11 for growth promotion of potato (Solanum tuberosum L.). Front. Microbiol. 6:583. doi: 10.3389/fmicb.2015.00583
Hansen, V., Bonnichsen, L., Nunes, I., Sexlinger, K., Lopez, S. R., van der Bom, F. J. T., et al. (2020). Seed inoculation with Penicillium bilaiae and Bacillus simplex affects the nutrient status of winter wheat. Biol. Fert. Soils 56, 97–109. doi: 10.1007/s00374-019-01401-7
Hu, Y., Xia, Y., Sun, Q., Liu, K., Chen, X., Ge, T., et al. (2018). Effects of long-term fertilization on phoD-harboring bacterial community in Karst soils. Sci. Total Environ. 628–629, 53–63. doi: 10.1016/j.scitotenv.2018.01.314
Lim, B. L., Yeung, P., Cheng, C., and Hill, J. E. (2007). Distribution and diversity of phytate-mineralizing bacteria. ISME J. 1, 321–330. doi: 10.1038/ismej.2007.40
Liu, F. P., Liu, H. Q., Zhou, H. L., Dong, Z. G., Bai, X. H., Bai, P., et al. (2014). Isolation and characterization of phosphate-solubilizing bacteria from betel nut (Areca catechu) and their effects on plant growth and phosphorus mobilization in tropical soils. Biol. Fert. Soils 50, 927–937. doi: 10.1007/s00374-014-0913-z
Luo, G., Ling, N., Nannipieri, P., Chen, H., Raza, W., Wang, M., et al. (2017). Long-term fertilisation regimes affect the composition of the alkaline phosphomonoesterase encoding microbial community of a vertisol and its derivative soil fractions. Biol. Fert. Soils 53, 375–388. doi: 10.1007/s00374-017-1183-3
Maougal, R. T., Brauman, A., Plassard, C., Abadie, J., Djekoun, A., and Drevon, J. J. (2014). Bacterial capacities to mineralize phytate increase in the rhizosphere of nodulated common bean (Phaseolus vulgaris) under P deficiency. Eur. J. Soil Biol. 62, 8–14. doi: 10.1016/j.ejsobi.2014.02.006
Marra, L. M., Soares, C. R. F. S., de Oliveira, S. M., Avelar Ferreira, P. A., Soares, B. L., and de Fráguas Carvalho, R. (2012). Biological nitrogen fixation and phosphate solubilization by bacteria isolated from tropical soils. Plant Soil 357, 289–307. doi: 10.1007/s11104-012-1157-z
Marwa, N., Singh, N., Srivastava, S., Saxena, G., and Pandey, V. (2019). Characterization the hypertolerance potential of two indigenous bacterial strains (Bacillus flexus and Acinetobacter junii) and their efficacy in arsenic bioremediation. J. Appl. Microbiol. 126, 1117–1127. doi: 10.1111/jam.14179
Mehrabi, Z., and Ramankutty, N. (2019). Synchronized failure of global crop production. Nat. Ecol. Evol. 3, 780–786. doi: 10.1038/s41559-019-0862-x
Mori, H., Maruyama, F., Kato, H., Toyoda, A., Dozono, A., Ohtsubo, Y., et al. (2013). Design and experimental application of a novel non-degenerate universal primer set that amplifies prokaryotic 16S rRNA genes with a low possibility to amplify eukaryotic rRNA genes. DNA Res. 21, 217–227. doi: 10.1093/dnares/dst052
Muñoz, G., Orlando, J., and Zuñiga-Feest, A. (2021). Plants colonizing volcanic deposits: root adaptations and effects on rhizosphere microorganisms. Plant Soil 461, 265–279. doi: 10.1007/s11104-020-04783-y
Nautiyal, C. S. (1999). An efficient microbiological growth medium for screening phosphate-solubilizing microorganisms. FEMS Microbiol. Lett. 170, 265–270. doi: 10.1016/S0378-1097(98)00555-2
Neal, A. L., Rossmann, M., Brearley, C., Akkari, E., Guyomar, C., Clark, I. M., et al. (2017). Land-use influences phosphatase gene microdiversity in soils. Environ. Microbiol. 19, 2740–2753. doi: 10.1111/1462-2920.13778
Oliveira, C. A., Alves, V. M. C., Marriel, I. E., Gomes, E. A., Scotti, M. R., Carneiro, N. P., et al. (2009). Phosphate solubilizing microorganisms isolated from rhizosphere of maize cultivated in an oxisol of the Brazilian Cerrado Biome. Soil Biol. Biochem. 41, 1782–1787. doi: 10.1016/j.soilbio.2008.01.012
Pastore, G., Kernchen, S., and Spohn, M. (2020). Microbial solubilization of silicon and phosphorus from bedrock in relation to abundance of phosphorus-solubilizing bacteria in temperate forest soils. Soil Biol. Biochem. 151:108050. doi: 10.1016/j.soilbio.2020.108050
Ragot, S. A., Huguenin-Elie, O., Kertesz, M. A., Frossard, E., and Bünemann, E. K. (2016). Total and active microbial communities and phoD as affected by phosphate depletion and pH in soil. Plant Soil 408, 15–30. doi: 10.1007/s11104-016-2902-5
Rasul, M., Yasmin, S., Suleman, M., Zaheer, A., Reitz, T., Tarkka, M. T., et al. (2019). Glucose dehydrogenase gene containing phosphobacteria for biofortification of phosphorus with growth promotion of rice. Microbiol. Res. 223–225, 1–12. doi: 10.1016/j.micres.2019.03.004
Richardson, A. E., Lynch, J. P., Ryan, P. R., Delhaize, E., Smith, F. A., Smith, S. E., et al. (2011). Plant and microbial strategies to improve the phosphorus efficiency of agriculture. Plant Soil 349, 121–156. doi: 10.1007/s11104-011-0950-4
Roberts, W. M., Matthews, R. A., Blackwell, M. S. A., Peukert, S., Collins, A. L., Stutter, M. I., et al. (2013). Microbial biomass phosphorus contributions to phosphorus solubility in riparian vegetated buffer strip soils. Biol. Fert. Soils 49, 1237–1241. doi: 10.1007/s00374-013-0802-x
Wagh, J., Shah, S., Bhandari, P., Archana, G., and Kumar, N. (2014). Heterologous expression of pyrroloquinoline quinone (pqq) gene cluster confers mineral phosphate solubilization ability to Herbaspirillum seropedicae Z67. Appl. Microbiol. Biotechnol. 98, 5117–5129. doi: 10.1007/s00253-014-5610-1
Wan, W., Hao, X., Xing, Y., Liu, S., Zhang, X., Li, X., et al. (2021a). Spatial differences in soil microbial diversity caused by pH-driven organic phosphorus mineralization. Land Degrad. Dev. 32, 766–776. doi: 10.1002/ldr.3734
Wan, W., Li, X., Han, S., Wang, L., Luo, X., Chen, W., et al. (2020a). Soil aggregate fractionation and phosphorus fraction driven by long-term fertilization regimes affect the abundance and composition of P-cycling-related bacteria. Soil Till. Res. 196:104475. doi: 10.1016/j.still.2019.104475
Wan, W., Liu, S., Li, X., Xing, Y., Chen, W., and Huang, Q. (2021b). Bridging rare and abundant bacteria with ecosystem multifunctionality in salinized agricultural soils: from community diversity to environmental adaptation. mSystems 6, e01221–20. doi: 10.1128/mSystems.01221-20
Wan, W., Liu, S., Li, X., Xing, Y., Chen, W., and Huang, Q. (2021c). Dispersal limitation driving phoD-harboring bacterial community assembly: a potential indicator for ecosystem multifunctionality in long-term fertilized soils. Sci. Total Environ. 754:141960. doi: 10.1016/j.scitotenv.2020.141960
Wan, W., Qin, Y., Wu, H., Zuo, W., He, H., Tan, J., et al. (2020b). Isolation and characterization of phosphorus solubilizing bacteria with multiple phosphorus sources utilizing capacity and their potential for lead immobilization in soil. Front. Microbiol. 11:752. doi: 10.3389/fmicb.2020.00752
Wei, X., Hu, Y., Razavi, B. S., Zhou, J., Shen, J., Nannipieri, P., et al. (2019). Rare taxa of alkaline phosphomonoesterase-harboring microorganisms mediate soil phosphorus mineralization. Soil Biol. Biochem. 131, 62–70. doi: 10.1016/j.soilbio.2018.12.025
Wei, Y., Zhao, Y., Fan, Y., Lu, Q., Li, M., Wei, Q., et al. (2017). Impact of phosphate-solubilizing bacteria inoculation methods on phosphorus transformation and long-term utilization in composting. Bioresour. Technol. 241, 134–141. doi: 10.1016/j.biortech.2017.05.099
Wemheuer, F., Taylor, J. A., Daniel, R., Johnston, E., Meinicke, P., Thomas, T., et al. (2020). Tax4Fun2: prediction of habitat-specific functional profiles and functional redundancy based on 16S rRNA gene sequences. Environ. Microbiome. 15:11. doi: 10.1186/s40793-020-00358-7
Widdig, M., Schleuss, P. M., Weig, A. R., Guhr, A., Biederman, L. A., Borer, E. T., et al. (2019). Nitrogen and phosphorus additions alter the abundance of phosphorus-solubilizing bacteria and phosphorus activity in grassland soils. Front. Env. Sci. 7:185. doi: 10.3389/fenvs.2019.001985
Xue, L., Ren, H. D., Li, S., Leng, X. H., and Yao, X. H. (2017). Soil bacterial community structure and co-occurrence pattern during vegetation restoration in karst rocky desertification area. Front. Microbiol. 8:2377. doi: 10.3389/fmicb.2017.02377
Yang, Y., Zhang, X. Y., Wang, H. M., Fu, X. L., Wen, X. F., Zhang, C., et al. (2018). How understory vegetation affects the catalytic properties of soil extracellular hydrolases in a Chinese fir (Cunninghamia lanceolata) forest. Eur. J. Soil Biol. 90, 15–21. doi: 10.1016/j.ejsobi.2018.11.004
Ye, D., Li, T., Zhang, X., Zheng, X., and Dai, W. (2017). Rhizosphere P composition, phosphatase and phytase activities of Polygonum hydropiper grown in excess P soils. Biol. Fert. Soils 53, 823–836. doi: 10.1007/s00374-017-1218-9
Yu, X., Liu, X., Zhu, T. H., Liu, G. H., and Mao, C. (2011). Isolation and characterization of phosphate-solubilizing bacteria from walnut and their effect on growth and phosphorus mobilization. Biol. Fert. Soils 47, 437–446. doi: 10.1007/s00374-011-0548-2
Keywords: phosphorus-solubilizing bacteria, P-cycling-related gene, rhizosphere bacterial community, functional profiling, vegetation properties
Citation: He D and Wan W (2021) Phosphate-Solubilizing Bacterium Acinetobacter pittii gp-1 Affects Rhizosphere Bacterial Community to Alleviate Soil Phosphorus Limitation for Growth of Soybean (Glycine max). Front. Microbiol. 12:737116. doi: 10.3389/fmicb.2021.737116
Received: 06 July 2021; Accepted: 27 August 2021;
Published: 24 September 2021.
Edited by:
Wei Zhang, Michigan State University, United StatesReviewed by:
Anukool Vaishnav, GLA University, IndiaAnandham Rangasamy, Tamil Nadu Agricultural University, India
Copyright © 2021 He and Wan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wenjie Wan, wanwenjie@wbgcas.cn
 Donglan He
Donglan He Wenjie Wan
Wenjie Wan