Abstract
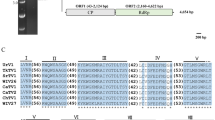
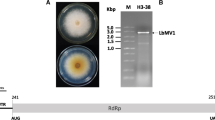
The complete genome of a novel mycovirus, Albatrellopsis flettii mitovirus 1 (AfMV1), hosted by the basidiomycetous ectomycorrhizal fungus Albatrellopsis flettii (Morse ex Pouzar) Audet, was sequenced and analyzed. The full-length cDNA sequence, obtained from a dsRNA replication intermediate of the AfMV1 genome, is 3037 bp in length with a predicted G+C content of 40.66%. Sequence analysis revealed that a single large open reading frame (ORF) is present on the positive strand when the mold mitochondrial genetic code is applied. The single ORF encodes a putative RNA-dependent RNA polymerase of 859 amino acids with a predicted molecular weight of 97.05 kDa that shares the closest similarity with the corresponding protein of Entomophthora muscae mitovirus 7, with 43.38% sequence identity. Phylogenetic analysis showed that AfMV1 could be classified as a new member of the genus Mitovirus within the family Mitoviridae. This is the first report of the complete genome sequence of a new mitovirus, AfMV1, isolated from the basidiomycetous ectomycorrhizal fungus A. flettii.


Similar content being viewed by others
References
Giehl RF, von Wiren N (2014) Root nutrient foraging. Plant Physiol 166(2):509–517. https://doi.org/10.1104/pp.114.245225
Brundrett MC, Tedersoo L (2018) Evolutionary history of mycorrhizal symbioses and global host plant diversity. New Phytol 220(4):1108–1115. https://doi.org/10.1111/nph.14976
Smith S, Read D (2008) Mycorrhizal symbiosis, 3rd, edition. Academic Press
Tripathi S, Mishra SK, Varma A (2017) Mycorrhizal Fungi as Control Agents Against Plant Pathogens. In: Varma A. PR, Tuteja N. (ed) Mycorrhiza - Nutrient Uptake, Biocontrol, Ecorestoration. Springer, Cham. doi:https://doi.org/10.1007/978-3-319-68867-1_8
Garg N, Singh S, Kashyap L (2017) Arbuscular Mycorrhizal Fungi and Heavy Metal Tolerance in Plants: An Insight into Physiological and Molecular Mechanisms. In: Varma A. PR, Tuteja N. (ed) Mycorrhiza - Nutrient Uptake, Biocontrol, Ecorestoration. Springer, Cham. doi:https://doi.org/10.1007/978-3-319-68867-1_4
Ginns J (1997) The taxonomy and distribution of rare or uncommon species of Albatrellus in western North America. Can J Bot 75(2):261–273. https://doi.org/10.1139/b97-028
Zheng HD, Liu PG (2008) Additions to our knowledge of the genus Albatrellus (Basidiomycota) in China. Fungal Divers 32:157–170
Altuntaş D, Şahin E, Kabaktepe S, Allı H, Akata I (2021) Albatrellopsis flettii, A New Genus for Turkish Albatrellaceae. KSU J Agric Nat. 24(4):815–819. https://doi.org/10.18016/ksutarimdoga.vi.799215
Petrzik K, Sarkisova T, Stary J, Koloniuk I, Hrabakova L, Kubesova O (2016) Molecular characterization of a new monopartite dsRNA mycovirus from mycorrhizal Thelephora terrestris (Ehrh.) and its detection in soil oribatid mites (Acari: Oribatida). Virology 489:12–19. https://doi.org/10.1016/j.virol.2015.11.009
Sahin E, Akata I (2019) Complete genome sequence of a novel mitovirus from the ectomycorrhizal fungus Geopora sumneriana. Arch Virol 164(11):2853–2857. https://doi.org/10.1007/s00705-019-04367-x
Sahin E, Akata I, Keskin E (2020) Novel and divergent bipartite mycoviruses associated with the ectomycorrhizal fungus Sarcosphaera coronaria. Virus Res 286:198071. https://doi.org/10.1016/j.virusres.2020.198071
Sahin E, Keskin E, Akata I (2020) Novel and diverse mycoviruses co-inhabiting the hypogeous ectomycorrhizal fungus Picoa juniperi. Virology 552:10–19. https://doi.org/10.1016/j.virol.2020.09.009
Sahin E, Akata I (2021) Full-length genome characterization of a novel alphapartitivirus detected in the ectomycorrhizal fungus Hygrophorus penarioides. Virus Genes 57:94–99. https://doi.org/10.1007/s11262-020-01814-9
Stielow B, Klenk HP, Menzel W (2011) Complete genome sequence of the first endornavirus from the ascocarp of the ectomycorrhizal fungus Tuber aestivum Vittad. Arch Virol 156(2):343–345. https://doi.org/10.1007/s00705-010-0875-x
Stielow B, Klenk HP, Winter S, Menzel W (2011) A novel Tuber aestivum (Vittad.) mitovirus. Arch Virol 156 (6):1107-1110. doi:https://doi.org/10.1007/s00705-011-0998-8
Stielow B, Menzel W (2010) Complete nucleotide sequence of TaV1, a novel totivirus isolated from a black truffle ascocarp (Tuber aestivum Vittad.). Arch Virol 155 (12):2075-2078. doi:https://doi.org/10.1007/s00705-010-0824-8
Stielow JB, Bratek Z, Klenk HP, Winter S, Menzel W (2012) A novel mitovirus from the hypogeous ectomycorrhizal fungus Tuber excavatum. Arch Virol 157(4):787–790. https://doi.org/10.1007/s00705-012-1228-8
Sutela S, Vainio EJ (2020) Virus population structure in the ectomycorrhizal fungi Lactarius rufus and L. tabidus at two forest sites in Southern Finland. Virus Res 285:197993. doi:https://doi.org/10.1016/j.virusres.2020.197993
Hillman BI, Annisa A, Suzuki N (2018) Viruses of Plant-Interacting Fungi. Adv Virus Res 100:99–116. https://doi.org/10.1016/bs.aivir.2017.10.003
Sahin E, Akata I (2018) Viruses infecting macrofungi. Virusdisease 29(1):1–18. https://doi.org/10.1007/s13337-018-0434-8
Ghabrial SA, Caston JR, Jiang D, Nibert ML, Suzuki N (2015) 50-plus years of fungal viruses. Virology 479–480:356–368. https://doi.org/10.1016/j.virol.2015.02.034
Sutela S, Poimala A, Vainio EJ (2019) Viruses of fungi and oomycetes in the soil environment. FEMS Microbiol Ecol 95 (9). doi:https://doi.org/10.1093/femsec/fiz119
Botella L, Janousek J, Maia C, Jung MH, Raco M, Jung T (2020) Marine oomycetes of the genus halophytophthora harbor viruses related to bunyaviruses. Front Microbiol 11:1467. https://doi.org/10.3389/fmicb.2020.01467
Ruiz-Padilla A, Rodriguez-Romero J, Gomez-Cid I, Pacifico D, Ayllon MA (2021) Novel Mycoviruses Discovered in the Mycovirome of a Necrotrophic Fungus. mBio 12 (3). doi:https://doi.org/10.1128/mBio.03705-20
Hillman BI, Cai G (2013) The family narnaviridae: simplest of RNA viruses. Adv Virus Res 86:149–176. https://doi.org/10.1016/B978-0-12-394315-6.00006-4
Nibert ML, Debat HJ, Manny AR, Grigoriev IV, De Fine Licht HH (2019) Mitovirus and Mitochondrial Coding Sequences from Basal Fungus Entomophthora muscae. Viruses 11 (4). doi:https://doi.org/10.3390/v11040351
Darissa O, Willingmann P, Adam G (2010) Optimized approaches for the sequence determination of double-stranded RNA templates. J Virol Methods 169(2):397–403. https://doi.org/10.1016/j.jviromet.2010.08.013
Madeira F, Park YM, Lee J, Buso N, Gur T, Madhusoodanan N, Basutkar P, Tivey ARN, Potter SC, Finn RD, Lopez R (2019) The EMBL-EBI search and sequence analysis tools APIs in 2019. Nucleic Acids Res 47(W1):W636–W641. https://doi.org/10.1093/nar/gkz268
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35(6):1547–1549. https://doi.org/10.1093/molbev/msy096
Li S, Li Y, Hu C, Han C, Zhou T, Zhao C, Wu X (2020) Full genome sequence of a new mitovirus from the phytopathogenic fungus Rhizoctonia solani. Arch Virol 165(7):1719–1723. https://doi.org/10.1007/s00705-020-04664-w
Xu Z, Wu S, Liu L, Cheng J, Fu Y, Jiang D, Xie J (2015) A mitovirus related to plant mitochondrial gene confers hypovirulence on the phytopathogenic fungus Sclerotinia sclerotiorum. Virus Res 197:127–136. https://doi.org/10.1016/j.virusres.2014.12.023
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This study did not include experiments with human participants or animals performed by any of the authors.
Additional information
Handling Editor: Robert H.A. Coutts.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Akata, I., Keskin, E. & Sahin, E. Molecular characterization of a new mitovirus hosted by the ectomycorrhizal fungus Albatrellopsis flettii. Arch Virol 166, 3449–3454 (2021). https://doi.org/10.1007/s00705-021-05250-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-021-05250-4