Abstract
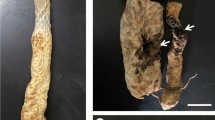
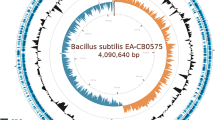
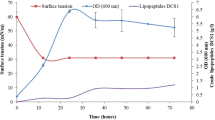
Verticilllium wilt of cotton is a devastating soil-borne disease, which is caused by Verticillium dahliae Kleb. Bacillus velezensis strain AL7 was isolated from cotton soil. This strain efficiently inhibited the growth of V. dahliae. But the mechanism of the biocontrol strain AL7 remains poorly understood. To understand the possible genetic determinants for biocontrol traits of this strain, we conducted phenotypic, phylogenetic and comparative genomics analysis. Phenotypic analysis showed that strain AL7 exhibited broad-spectrum antifungal activities. We determined that the whole genome sequence of B. velezensis AL7 is a single circular chromosome that is 3.89 Mb in size. The distribution of putative gene clusters that could benefit to biocontrol activities was found in the genome. Phylogenetic analysis of Bacillus strains by using single core-genome clearly placed strain AL7 into the B. velezensis. Meantime, we performed comparative analyses on four Bacillus strains and observed subtle differences in their genome sequences. In addition, comparative genomics analysis showed that the core genomes of B. velezensis are more abundant in genes relevant to secondary metabolism compared with B. subtilis strains. Single mutant in the biosynthetic genes of fengycin demonstrated the function of fengycin in the antagonistic activity of B. velezensis AL7. Here, we report a new biocontrol bacterium B. velezensis AL7 and fengycin contribute to the biocontrol efficacy of the strain. The results showed in the research further sustain the potential of B. velezensis AL7 for application in agriculture production and may be a worthy biocontrol strain for further studies.






Similar content being viewed by others
References
Berg G, Koeberl M, Rybakova D, Mueller H, Grosch R, Smalla K (2017) Plant microbial diversity is suggested as the key to future biocontrol and health trends. FEMS Microbiol Ecol 93:fix050. https://doi.org/10.1093/femsec/fix050
Blom J, Rueckert C, Niu B, Wang Q, Borriss R (2012) The complete genome of Bacillus amyloliquefaciens subsp plantarum CAU B946 contains a gene cluster for nonribosomal synthesis of Iturin A. J Bacteriol 194:1845–1846
Branda SS, Gonzalez-Pastor JE, Ben-Yehuda S, Losick R, Kolter R (2001) Fruiting body formation by Bacillus subtilis. P Natl Acad Sci USA 98:11621–11626
Cao Y, Pi H, Chandrangsu P, Li Y, Wang Y, Zhou H, Xiong H, Helmann JD, Cai Y (2018) Antagonism of two plant-growth promoting Bacillus velezensis isolates against Ralstonia solanacearum and Fusarium oxysporum. Sci Rep 8:4360
Castresana J (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17:540–552
Chen X, Zhang Y, Fu X, Li Y, Wang Q (2016) Isolation and characterization of Bacillus amyloliquefaciens PG12 for the biological control of apple ring rot. Postharvest Biol Tec 115:113–121
Darling A, Mau B, Blattner FR, Perna NT (2004) Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res 14:1394–1403
Eljounaidi K, Lee SK, Bae H (2016) Bacterial endophytes as potential biocontrol agents of vascular wilt diseases—review and future prospects. Biol Control 103:62–68
Erdogan O, Benlioglu K (2010) Biological control of Verticillium wilt on cotton by the use of fluorescent Pseudomonas spp. under field conditions. Biol Control 53:39–45
Fan B, Blom J, Klenk H, Borriss R (2017a) Bacillus amyloliquefaciens, Bacillus velezensis, and Bacillus siamensis form an “operational group B. amyloliquefaciens” within the B. subtilis species complex. Front Microbiol 8:22
Fan H, Zhang Z, Li Y, Zhang X, Duan Y, Wang Q (2017b) Biocontrol of bacterial fruit blotch by Bacillus subtilis 9407 via surfactin-mediated antibacterial activity and colonization. Front Microbiol 8:1973
Gomez-Lama Cabanas C, Legarda G, Ruano-Rosa D, Pizarro-Tobias P, Valverde-Corredor A, Niqui JL, Trivino JC, Roca A, Mercado-Blanco J (2018) Indigenous Pseudomonas spp. strains from the Olive (Olea europaea L) rhizosphere as effective biocontrol agents against Verticillium dahlia: from the host roots to the bacterial genomes. Front Microbiol 9:277
Han Q, Wu F, Wang X, Qi H, Shi L, Ren A, Liu Q, Zhao M, Tang C (2015) The bacterial lipopeptide iturins induce Verticillium dahliae cell death by affecting fungal signalling pathways and mediate plant defence responses involved in pathogen-associated molecular pattern-triggered immunity. Environ Microbiol 17:1166–1188
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780
Kotb E (2015) Purification and partial characterization of serine fibrinolytic enzyme from Bacillus megaterium KSK-07 isolated from kishk, a traditional Egyptian fermented food. Appl Biochem Micro 51:34–43
Koumoutsi A, Chen XH, Henne A, Liesegang H, Hitzeroth G, Franke P, Vater J, Borriss R (2004) Structural and functional characterization of gene clusters directing nonribosomal synthesis of bioactive cyclic lipopeptides in Bacillus amyloliquefaciens strain FZB42. J BACTERIOL 186:1084–1096
Li S, Zhang N, Zhang Z, Luo J, Shen B, Zhang R, Shen Q (2013) Antagonist Bacillus subtilis HJ5 controls Verticillium wilt of cotton by root colonization and biofilm formation. Biol Fert Soils 49:295–303
Li Y, Heloir M, Zhang X, Geissler M, Trouvelot S, Jacquens L, Henkel M, Su X, Fang X, Wang Q, Adrian M (2019) Surfactin and fengycin contribute to the protection of a Bacillus subtilis strain against grape downy mildew by both direct effect and defence stimulation. Mol Plant Pathol 20:1037–1050
Liu H, Yin S, An L, Zhang G, Cheng H, Xi Y, Cui G, Zhang F, Zhang L (2016) Complete genome sequence of Bacillus subtilis BSD-2, a microbial germicide isolated from cultivated cotton. J Biotechnol 230:26–27
Liu H, ZengQ Wang W, Zhang R, Yao J, Gill SR (2020) Complete genome sequence of Bacillus velezensis strain AL7, a biocontrol agent for suppression of cotton Verticillium wilt. Microbiol Resour Announc 9:e01595–19. https://doi.org/10.1128/MRA.01595-19
Luo C, Zhou H, Zou J, Wang X, Zhang R, Xiang Y, Chen Z (2015) Bacillomycin L and surfactin contribute synergistically to the phenotypic features of Bacillus subtilis 916 and the biocontrol of rice sheath blight induced by Rhizoctonia solani. Appl Microbiol Biot 99:1897–1910
Magno-Perez-Bryan MC, Martinez-Garcia PM, Hierrezuelo J, Rodriguez-Palenzuela P, Arrebola E, Ramos C, de Vicente A, Perez-Garcia A, Romero D (2015) Comparative genomics within the Bacillus genus reveal the singularities of two robust Bacillus amyloliquefaciens biocontrol strains. Mol Plant Microbe in 28:1102–1116
Martin JA, Macaya-Sanz D, Witzell J, Blumenstein K, Gil L (2015) Strong in vitro antagonism by elm xylem endophytes is not accompanied by temporally stable in planta protection against a vascular pathogen under field conditions. Eur J Plant Pathol 142:185–196
Oerke EC, Dehne HW (2004) Safeguarding production—losses in major crops and the role of crop protection. Crop Prot 23:275–285
Paterson J, Jahanshah G, Li Y, Wang Q, Mehnaz S, Gross H (2017) The contribution of genome mining strategies to the understanding of active principles of PGPR strains. FEMS Microbiol Ecol 93:fiw249. https://doi.org/10.1093/femsec/fiw249
Richter M, Rossello-Mora R (2009) Shifting the genomic gold standard for the prokaryotic species definition. P Natl Acad Sci USA 106:19126–19131
Romero D, de Vicente A, Rakotoaly RH, Dufour SE, Veening J, Arrebola E, Cazorla FM, Kuipers OP, Paquot M, Perez-Garcia A (2007) The iturin and fengycin families of lipopeptides are key factors in antagonism of Bacillus subtilis toward Podosphaera fusca. Mol Plant Microbe in 20:430–440
Shafi J, Tian H, Ji M (2017) Bacillus species as versatile weapons for plant pathogens: a review. Biotechnol Biotec Eq 31:446–459
Tan T, Zhu J, Shen A, Li J, Yu Y, Zhang M, Zhao M, Li Z, Chen J, Gao C, Cheng Y, Guo L, Yan L, Sun X, Zeng L, Yan Z (2019) Isolation and identification of a Bacillus subtilis HZ-72 exhibiting biocontrol activity against flax seedling blight. Eur J Plant Pathol 153:825–836
Tettelin H, Riley D, Cattuto C, Medini D (2008) Comparative genomics: the bacterial pan-genome. Curr Opin Microbiol 11:472–477
Tidjiani Alou M, Rathored J, Khelaifia S, Michelle C, Brah S, Diallo BA, Raoult D, Lagier J (2015) Bacillus rubiinfantis sp. nov. strain mt2T, a new bacterial species isolated from human gut. New Microbes New Infect 8:51–60
Wang X, Wei Z, Yang K, Wang J, Jousset A, Xu Y, Shen Q, Friman V (2019) Phage combination therapies for bacterial wilt disease in tomato. Nat Biotechnol 37:1513–1520
Weng J, Wang Y, Li J, Shen Q, Zhang R (2013) Enhanced root colonization and biocontrol activity of Bacillus amyloliquefaciens SQR9 by abrB gene disruption. Appl Microbiol Biot 97:8823–8830
Wu L, Wu H, Qiao J, Gao X, Borriss R (2015) Novel routes for improving biocontrol activity of Bacillus based bioinoculants. Front Microbiol 6:1395
Xu Z, Shao J, Li B, Yan X, Shen Q, Zhang R (2013) Contribution of Bacillomycin D in Bacillus amyloliquefaciens SQR9 to antifungal activity and biofilm formation. Appl Environ Microb 79:808–815
Xue L, Gu M, Xu W, Lu J, Xue Q (2016) Antagonistic Streptomyces enhances defense-related responses in cotton for biocontrol of wilt caused by phytotoxin of Verticillium dahliae. Phytoparasitica 44:225–237
Yoon S, Ha S, Lim J, Kwon S, Chun J (2017) A large-scale evaluation of algorithms to calculate average nucleotide identity. Anton Leeuw Int J G 110:1281–1286
Zeng Q, Xie J, Li Y, Gao T, Xu C, Wang Q (2018) Comparative genomic and functional analyses of four sequenced Bacillus cereus genomes reveal conservation of genes relevant to plant-growth- promoting traits. Sci Rep-UK 8:17009. https://doi.org/10.1038/s41598-018-35300-y
Zeriouh H, Romero D, Garcia-Gutierrez L, Cazorla FM, de Vicente A, Perez-Garcia A (2011) The iturin-like lipopeptides are essential components in the biological control arsenal of Bacillus subtilis against bacterial diseases of cucurbits. Mol Plant Microbe in 24:1540–1552
Zhang X, Zhou Y, Li Y, Fu X, Wang Q (2017) Screening and characterization of endophytic Bacillus for biocontrol of grapevine downy mildew. Crop Prot 96:173–179
Zhang F, Li X, Zhu S, Ojaghian MR, Zhang J (2018) Biocontrol potential of Paenibacillus polymyxa against Verticillium dahliae infecting cotton plants. Biol Control 127:70–77
Zhang L, Li W, Tao Y, Zhao S, Yao L, Cai Y, Niu Q (2019) Overexpression of the key virulence factor 1,3–1,4-beta-D-glucanase in the endophytic bacterium Bacillus halotolerans Y6 to improve Verticillium resistance in cotton. J Agr Food Chem 67:6828–6836
Zhang N, Yang D, Kendall JRA, Borriss R, Druzhinina IS, Kubicek CP, Shen Q, Zhang R (2016) Comparative genomic analysis of Bacillus amyloliquefaciens and Bacillus subtilis reveals evolutional traits for adaptation to plant-associated habitats. Front Microbiol 7:2039
Zhao Y, Wu J, Yang J, Sun S, Xiao J, Yu J (2012) PGAP: pan-genomes analysis pipeline. Bioinformatics 28:416–418
Zhao Y, Jia X, Yang J, Ling Y, Zhang Z, Yu J, Wu J, Xiao J (2014) PanGP: a tool for quickly analyzing bacterial pan-genome profile. Bioinformatics 30:1297–1299
Zhao H, Shao D, Jiang C, Shi J, Li Q, Huang Q, Rajoka MSR, Yang H, Jin M (2017) Biological activity of lipopeptides from Bacillus. Appl Microbiol Biot 101:5951–5960
Acknowledgements
This study was funded by the project of renovation capacity building for the young sci-tech talents sponsored by Xinjiang academy of agricultural sciences (xjnkq-2020015), The National Key Research and Development Program of China (No. 2017YFD020030303), Science and technology support project of Xinjiang Autonomous Region (2019E0244).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Haiyang Liu declares that he has no conflict of interest. Qingchao Zeng declares that he has no conflict of interest. Nuerziya Yalimaimaiti declares that she has no conflict of interest. Wei Wang declares that he has no conflict of interest. Renfu Zhang declares that he has no conflict of interest. Ju Yao declares that he has no conflict of interest.
Ethical approval
The artical does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
438_2021_1816_MOESM1_ESM.tif
Supplementary file1 Figure S1 Genome map of B. velezensisstrainAL7. Each ring of the circle represents a different genome information: rings from the outside inward: (1) scale marks (unit: Mb), (2, 3) protein-coding genes on the forward and reverse strands, respectively (different color represent the different functional classifications), (4, 5) rRNA (blue) and tRNA (red) on the forward and reverse strands, respectively, (6) GC content (positive: red; negative: blue), and (7) GC skew (above average: aquamarine; below average: orange). (TIF 19,804 KB)
438_2021_1816_MOESM4_ESM.tif
Supplementary file4 Figure S4 Genome comparisons of other B. velezensisstrains against reference genome (strain FZB42) generating through BRIG version 0.95. The circular map explains the whole genome comparison of biocontrol strain AL7 against the other three selected B. velezensis. Theinner cycle displays the genome sequence of the reference strain AL7 and the shade of colors display the similarities between each strains with biocontrol strain AL7. (TIF 4,038 KB)
438_2021_1816_MOESM5_ESM.tif
Supplementary file5 Figure S5 Genome alignment showing synthetic blocks between Bacillusstrains. The same colored blocks represent the homologous DNA regions among the strains, and gaps display the non-homologous regions. (TIF 1,864 KB)
438_2021_1816_MOESM8_ESM.xlsx
Supplementary file8 Table S3 Genes and gene cluster encoding for secondary metabolites in Bacillus velezensis AL7. (XLSX 10 KB)
438_2021_1816_MOESM10_ESM.xlsx
Supplementary file10 Table S5 Comparative analysis of the functional classifications based on COG of the protein-coding genes of the Bacillusstrains analyzed. (XLSX 10 KB)
Rights and permissions
About this article
Cite this article
Liu, H., Zeng, Q., Yalimaimaiti, N. et al. Comprehensive genomic analysis of Bacillus velezensis AL7 reveals its biocontrol potential against Verticillium wilt of cotton. Mol Genet Genomics 296, 1287–1298 (2021). https://doi.org/10.1007/s00438-021-01816-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-021-01816-8