Abstract
Alphasatellites (family Alphasatellitidae) are circular, single-stranded DNA molecules (~1–1.4 kb) that encode a replication-associated protein and have commonly been associated with some members of the families Geminiviridae, Nanoviridae, and Metaxyviridae (recently established). Here, we provide a taxonomy update for the family Alphasatellitidae following the International Committee on Taxonomy of Viruses (ICTV) Ratification Vote held in March 2021. The taxonomic update includes the establishment of the new subfamily Petromoalphasatellitinae. This new subfamily includes three new genera as well as the genus Babusatellite, which previously belonged to the subfamily Nanoalphasatellitinae. Additionally, three new genera and 14 new species have been established in the subfamily Geminialphasatellitinae, as well as five new species in the subfamily Nanoalphasatellitinae.
Similar content being viewed by others
Introduction
Alphasatellitidae is a family of replication-associated protein (Rep)-expressing circular, single-stranded DNA molecules (~1–1.4 kb) that have commonly been found in association with some members of the families Geminiviridae (viruses in the genera Begomovirus and Mastrevirus) [14] and Nanoviridae [12]. Similar molecules have also been found associated with coconut foliar decay virus [4] (family Metaxyviridae, genus Cofodevirus) [13]. Alphasatellites are unable to trans-replicate the bona fide genome components of these ‘helper viruses’ or vice versa, but they do rely on them for encapsidation, movement, and vector transmission [1].
Since the establishment of the family Alphasatellitidae in 2018 [1], various new alphasatellites have been identified, and here, we provide a taxonomic update. The update includes: (1) the establishment of a new subfamily, Petromoalphasatellitinae; (2) reassignment of existing family members and assignment of new members to the subfamily Petromoalphasatellitinae; (3) the establishment of three new genera and 14 new species in the subfamily Geminialphasatellitinae; and (4) the establishment of five new species in the subfamily Nanoalphasatellitinae.
Geminialphasatellitinae
A genus demarcation threshold of 70% and a species demarcation threshold of 88% based on genome-wide pairwise identity values were recommended by Briddon et al. [1] (Fig. 1). Accordingly, the members were classified into four genera (Ageyesisatellite, Clecrusatellite, Colecusatellite, and Gosmusatellite) and a total of 41 species. In the last couple of years, various geminialphasatellites have been identified, and using the taxonomic guidelines outlined by Briddon et al. [1], we established three new genera (Draflysatellite, Somasatellite, and Whiflysatellite) to accommodate the two previously unassigned species (Dragonfly associated alphasatellite and Whitefly associated Guatemala alphasatellite 1) and one new species (Sorghum mastrevirus associated alphasatellite). In addition to these, we established 12 new species in the genera Clecrusatellite (n = 7), Colecusatellite (n = 2), and Gosmusatellite (n = 3). The details of the new taxa are provided in Table 1. With all the new geminialphasatellites being identified, we reanalysed the distribution of pairwise identity values for 960 complete sequences with intact Rep open reading frames using SDT v1.2 [7] and determined that the 88% species demarcation threshold is still a valid criterion for their classification (Fig. 1).
Distribution of pairwise identity values of complete sequences with intact Rep ORFs of members of the subfamilies Geminialphasatellitinae (n = 960), Nanoalphasatellitinae (n = 104), and Petromoalphasatellitinae (n = 28), determined using SDT v1.2 [7]
Nanoalphasatellitinae
A genus demarcation threshold of 67% and a species demarcation threshold of 80% based on genome-wide pairwise identity values were recommended previously by Briddon et al. [1]. This resulted in the original creation of 19 species, which were assigned to seven genera (Babusatellite, Clostunsatellite, Fabenesatellite, Milvetsatellite, Mivedwarsatellite, Sophoyesatellite, and Subclovsatellite). The major change in this subfamily is the reassignment of the genus Babusatellite to the subfamily Petromoalphasatellitinae. We also established five new species in the genera Mivedwarsatellite (n = 3), Sophoyesatellite (n = 1), and Subclovsatellite (n = 1). To verify that, with the reassignment of Babusatellite to the subfamily Petromoalphasatellitinae, the species demarcation threshold of 80% is still valid, we reanalysed the distribution of pairwise identity values for 104 complete nanoalphasatellite sequences with intact Rep open reading frames using SDT v1.2 [7] and confirmed the species demarcation threshold (Fig. 1).
Petromoalphasatellitinae
In 2018, novel types of single-stranded DNA molecules comprising the genome of coconut foliar decay virus [4] (family Metaxyviridae) were identified. The virus was found to affect only coconut palm (Cocos nucifera) and is restricted to the Vanuatu Archipelago [8, 9]. Nine different alphasatellites are associated with coconut foliar decay virus. Our analysis of these alphasatellites revealed that, based on a species demarcation threshold of 81% sequence identity, they can be classified in several distinct species (Fig. 1). Applying a genus demarcation threshold of 68% sequence identity among members, three new genera, Cocosatellite (Coco for coconut), Coprasatellite (Copra for coconut meat), and Kobbarisatellite (Kobbari is an Indian dish from coconut meat) were established. Five of the seven alphasatellite species were assigned to the genus Cocosatellite, one to the genus Coprasatellite, and one to the genus Kobbarisatellite (Table 1).
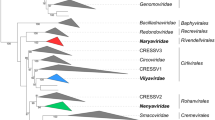
In addition to the creation of these three genera, we reassigned the genus Babusatellite to the new subfamily Petromoalphasatellitinae, as these alphasatellites collectively associate with viruses that infect perennial tropical monocotyledonous plants. Moreover, their respective size range is between those of the members of the subfamilies Geminialphasatellitinae and Nanoalphasatellitinae. Finally, phylogenetic analysis clearly supports inclusion of the species of the genus Babusatellite in the same subfamily along with the species of the newly established genera Cocosatellite, Coprasatellite, and Kobbarisatellite (Fig. 2). The distribution of the pairwise identity values for the 28 petromoalphasatellites is shown in Fig. 1.
Maximum-likelihood phylogenetic tree constructed from a MUSCLE [3] sequence alignment of representative alphasatellites using PHYML [5] with the GTR+I+G4 nucleotide substitution model (determined to be the best-fitting model by jModelTest [2]). The tree shows support for the proposed establishment of the new subfamily Petromoalphasatellitinae as well as support for the reassignment of the viruses in the genus Babuvirus. Branches with less than 60% bootstrap support have been collapsed using TreeGraph2 [11]. The tree was visualized in iToL v6 [6].
Based on the phylogenetic analysis coupled with pairwise comparisons, we moved three species (Banana bunchy top alphasatellite 2, Banana bunchy top alphasatellite 3, and Cardamom bushy dwarf alphasatellite) from the genus Babusatellite to a new genus, Muscarsatellite (Muscar from Musa and Elettaria cardamomum). A summary of the genera and species is provided in Table 1.
For the classification of new petromoalphasatellites, we recommend similar steps to those outlined by Briddon et al. [1]. To resolve conflict in cases where (1) a complete petromoalphasatellite sequence shares ≥81% pairwise identity with sequences of members of two different species or (2) a complete petromoalphasatellite sequence shares ≥81% pairwise identity with sequences of one or more members of a particular species while sharing <81% identity with the sequences of the majority of the members of that particular species:
-
(1)
The new petromoalphasatellite should be assigned to the species whose members share with it the highest pairwise sequence identity.
-
(2)
The petromoalphasatellite should be classified as belonging to any species in which it shares ≥81% pairwise sequence identity with any existing member of that species, even if it has <81% pairwise sequence identity to other members of that species.
Concluding remarks
As more sequence data become available, more changes to Alphasatellitidae taxonomy will certainly occur. We would also like to inform the Alphasatellitidae research community that a standardized binomial species nomenclature, consisting of the genus name and a free-form species epithet, has been ratified by the International Committee on Taxonomy of Viruses (ICTV) [10]. This needs to be adopted for currently established species in the family Alphasatellitidae by the year 2023. Thus, we encourage the community to engage with the ICTV Geminiviridae and Tolecusatellitidae Study Group and the Nanoviridae Study Group to determine the binomial names for current and new species in the family Alphasatellitidae.
References
Briddon RW, Martin DP, Roumagnac P, Navas-Castillo J, Fiallo-Olivé E, Moriones E, Lett JM, Zerbini FM, Varsani A (2018) Alphasatellitidae: a new family with two subfamilies for the classification of geminivirus- and nanovirus-associated alphasatellites. Arch Virol 163:2587–2600
Darriba D, Taboada GL, Doallo R, Posada D (2012) jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9:772
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Gronenborn B, Randles JW, Knierim D, Barriere Q, Vetten HJ, Warthmann N, Cornu D, Sileye T, Winter S, Timchenko T (2018) Analysis of DNAs associated with coconut foliar decay disease implicates a unique single-stranded DNA virus representing a new taxon. Sci Rep 8:5698
Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321
Letunic I, Bork P (2021) Interactive Tree Of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res 49:W293–W296
Muhire BM, Varsani A, Martin DP (2014) SDT: a virus classification tool based on pairwise sequence alignment and identity calculation. PLoS One 9:e108277
Randles JW, Julia JF, Calvez C, Dollet M (1986) Association of single-stranded DNA with the foliar decay disease of coconut palm in Vanuatu. Phytopathology 76:889–894
Randles JW, Hanold D, Julia JF (1987) Small circular single-stranded DNA associated with foliar decay disease of coconut palm in Vanuatu. J Gen Virol 68:273–280
Siddell SG, Walker PJ, Lefkowitz EJ, Mushegian AR, Dutilh BE, Harrach B, Harrison RL, Junglen S, Knowles NJ, Kropinski AM, Krupovic M, Kuhn JH, Nibert ML, Rubino L, Sabanadzovic S, Simmonds P, Varsani A, Zerbini FM, Davison AJ (2020) Binomial nomenclature for virus species: a consultation. Arch Virol 165:519–525
Stover BC, Muller KF (2010) TreeGraph 2: combining and visualizing evidence from different phylogenetic analyses. BMC Bioinf 11:7
Thomas JE, Gronenborn B, Harding RM, Mandal B, Grigoras I, Randles JW, Sano Y, Timchenko T, Vetten HJ, Yeh HH, Ziebell H, ICTV Report Consortium (2021) ICTV virus taxonomy profile: Nanoviridae. J Gen Virol 102
Walker PJ, Siddell SG, Lefkowitz EJ, Mushegian AR, Adriaenssens EM, Alfenas-Zerbini P, Davison AJ, Dempsey DM, Dutilh BE, Garcia ML, Harrach B, Harrison RL, Hendrickson RC, Junglen S, Knowles NJ, Krupovic M, Kuhn JH, Lambert AJ, Lobocka M, Nibert ML, Oksanen HM, Orton RJ, Robertson DL, Rubino L, Sabanadzovic S, Simmonds P, Smith DB, Suzuki N, Van Dooerslaer K, Vandamme AM, Varsani A, Zerbini FM (2021) Changes to virus taxonomy and to the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses (2021). Arch Virol 166:2633–2648
Zerbini FM, Briddon RW, Idris A, Martin DP, Moriones E, Navas-Castillo J, Rivera-Bustamante R, Roumagnac P, Varsani A, ICTV Report Consortium (2017) ICTV virus taxonomy profile: Geminiviridae. J Gen Virol 98:131–133
Acknowledgment
JML is supported by the European Union (ERDF), the Conseil Régional de La Réunion, and CIRAD.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Additional information
Handling Editor: Sead Sabanadzovic.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Varsani, A., Martin, D.P., Randles, J.W. et al. Taxonomy update for the family Alphasatellitidae: new subfamily, genera, and species. Arch Virol 166, 3503–3511 (2021). https://doi.org/10.1007/s00705-021-05232-6
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-021-05232-6