Abstract
Purpose
In hopes of discovering new markers for metastatic or aggressive phenotypes of pheochromocytomas and paragangliomas (PCPG), we analyzed the noncoding transcriptome from patient gene expression data in The Cancer Genome Atlas.
Methods
Differential expression of miRNAs was observed between PCPG molecular subtypes. We specifically characterized candidate miRNAs that are upregulated in pseudohypoxic PCPGs with mutations in succinate dehydrogenase complex subunits, B and/or D (SDHB and/or SDHD, respectively), which are mutations associated with unfavorable clinical outcomes.
Results
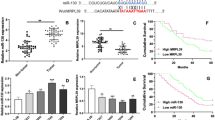
Our computational analysis identified four candidate miRNAs that showed elevated expression in metastatic compared to non-metastatic PCPGs: miR-182, miR-183, miR-96, and miR-383. We also found six candidate lncRNAs harboring opposite expression patterns from the miRNAs when we analyzed the expression profiles of their predicted target lncRNAs. Three of these lncRNA candidates, USP3-AS1, LINC00877, and AC009312.1, were validated to have reduced expression in metastatic compared to non-metastatic PCPGs. Finally, using univariate and multivariate analysis, we found miRNA miR-182 to be an independent predictor of metastasis-free survival in PCPGs.
Conclusions
We identified candidate miRNA and lncRNAs associated with metastasis-free survival in PCPGs.



Similar content being viewed by others
Data availability
The data used in this paper are available from The Cancer Genome Atlas at the Genomic Data Commons portal: https://gdc.cancer.gov.
Code availability
The R scripts used for this paper are available from https://github.com/suman-ghosal.
References
T. Zelinka, G. Eisenhofer, K. Pacak, Pheochromocytoma as a catecholamine producing tumor: implications for clinical practice. Stress 10(2), 195–203 (2007)
S. Hescot et al. Prognosis of Malignant Pheochromocytoma and Paraganglioma (MAPP-Prono Study): A European Network for the Study of Adrenal Tumors Retrospective Study. J. Clin. Endocrinol. Metab. 104(6), 2367–2374 (2019)
L. Fishbein et al. Comprehensive molecular characterization of pheochromocytoma and paraganglioma. Cancer Cell 31(2), 181–193 (2017)
J. Crona et al. Genotype-phenotype correlations in pheochromocytoma and paraganglioma: a systematic review and individual patient meta-analysis. Endocr. Relat. Cancer 26(5), 539–550 (2019)
I. Jochmanova et al. Hypoxia-inducible factor signaling in pheochromocytoma: turning the rudder in the right direction. J. Natl Cancer Inst. 105(17), 1270–1283 (2013)
L. Amar et al. Genetic testing in pheochromocytoma or functional paraganglioma. J. Clin. Oncol. 23(34), 8812–8818 (2005)
K.S. King et al. Metastatic pheochromocytoma/paraganglioma related to primary tumor development in childhood or adolescence: significant link to SDHB mutations. J. Clin. Oncol. 29(31), 4137–4142 (2011)
Y. Assadipour et al. SDHB mutation status and tumor size but not tumor grade are important predictors of clinical outcome in pheochromocytoma and abdominal paraganglioma. Surgery 161(1), 230–239 (2017)
A. Jha et al. Clinical, diagnostic, and treatment characteristics of SDHA-related metastatic pheochromocytoma and paraganglioma. Front. Oncol. 9, 53 (2019)
C. Trapnell et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28(5), 511–515 (2010)
R.C. Lee, V. Ambros, An extensive class of small RNAs in Caenorhabditis elegans. Science 294(5543), 862–864 (2001)
D.P. Bartel, MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116(2), 281–297 (2004)
X. Wang et al. Silencing of long noncoding RNA MALAT1 by miR-101 and miR-217 inhibits proliferation, migration, and invasion of esophageal squamous cell carcinoma cells. J. Biol. Chem. 290(7), 3925–3935 (2015)
A.N. Kallen et al. The imprinted H19 lncRNA antagonizes let-7 microRNAs. Mol. Cell 52(1), 101–112 (2013)
G. Adrian et al. MicroRNA signature associated with prognosis and progression in chronic Lymphocytic Leukemia. New England J. Med. 353(17), 1793–1801 (2005). https://doi.org/10.1056/NEJMoa050995
J.A. Chan, A.M. Krichevsky, K.S. Kosik, MicroRNA-21 Is an Antiapoptotic Factor in Human Glioblastoma Cells. Cancer Res. 65(14), 6029–6033 (2015). https://doi.org/10.1158/0008-5472.CAN-05-0137
A. Cimmino et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad. Sci. 102(39), 13944–13949 (2005). https://doi.org/10.1073/pnas.0506654102
S. Ghosal et al. Long intergenic noncoding RNA profiles of pheochromocytoma and paraganglioma: a novel prognostic biomarker. Int. J. Cancer 146(8), 2326–2335 (2020)
S. Pillai et al. MicroRNA 183 family profiles in pheochromocytomas are related to clinical parameters and SDHB expression. Hum. Pathol. 64, 91–97 (2017)
M.D. Robinson, D.J. McCarthy, G.K. Smyth, edgeR: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26(1), 139–140 (2010)
A. Jeggari, D.S. Marks, E. Larsson, miRcode: a map of putative microRNA target sites in the long non-coding transcriptome. Bioinformatics 28(15), 2062–2063 (2012)
S. Ghosal et al. miRepress: modelling gene expression regulation by microRNA with non-conventional binding sites. Sci. Rep. 6, 22334 (2016)
L.J. Castro-Vega et al. Multi-omics analysis defines core genomic alterations in pheochromocytomas and paragangliomas. Nat. Commun. 6, 6044 (2015)
A.A. de Cubas et al. Integrative analysis of miRNA and mRNA expression profiles in pheochromocytoma and paraganglioma identifies genotype-specific markers and potentially regulated pathways. Endocr. Relat. Cancer 20(4), 477–493 (2013)
M. Ayala-Ramirez et al. Clinical risk factors for malignancy and overall survival in patients with pheochromocytomas and sympathetic paragangliomas: primary tumor size and primary tumor location as prognostic indicators. J. Clin. Endocrinol. Metab. 96(3), 717–725 (2011)
B. Hu et al. POSTAR: a platform for exploring post-transcriptional regulation coordinated by RNA-binding proteins. Nucleic Acids Res. 45(D1), D104–D114 (2017)
Y.K. Zhou et al. Predicting lncRNA-protein interactions with miRNAs as mediators in a heterogeneous network model. Front. Genet. 10, 1341 (2019)
G. Eisenhofer et al. Measurements of plasma methoxytyramine, normetanephrine, and metanephrine as discriminators of different hereditary forms of pheochromocytoma. Clin. Chem. 57(3), 411–420 (2011)
G. Eisenhofer et al. Catecholamine metabolomic and secretory phenotypes in phaeochromocytoma. Endocr.-Relat. Cancer 18(1), 97–111 (2011)
H. Turkova et al. Characteristics and outcomes of metastatic Sdhb and sporadic pheochromocytoma/paraganglioma: an National Institutes of Health study. Endocr. Pract. 22(3), 302–314 (2016)
L. Ben Aim et al. Targeted next-generation sequencing detects rare genetic events in pheochromocytoma and paraganglioma. J. Med. Genet. 56(8), 513–520 (2019)
J. Welander, P. Soderkvist, O. Gimm, Genetics and clinical characteristics of hereditary pheochromocytomas and paragangliomas. Endocr.-Relat. Cancer 18(6), R253–R276 (2011)
G. Eisenhofer et al. Plasma methoxytyramine: a novel biomarker of metastatic pheochromocytoma and paraganglioma in relation to established risk factors of tumour size, location and SDHB mutation status. Eur. J. Cancer 48(11), 1739–1749 (2012)
S. Job et al. Telomerase activation and ATRX mutations are independent risk factors for metastatic pheochromocytoma and paraganglioma. Clin. Cancer Res. 25(2), 760–770 (2019)
L. Fishbein et al. Whole-exome sequencing identifies somatic ATRX mutations in pheochromocytomas and paragangliomas. Nat. Commun. 6, 6140 (2015)
T. Dwight et al. TERT structural rearrangements in metastatic pheochromocytomas. Endocr.-Relat. Cancer 25(1), 1–9 (2018)
O. Hamidi et al. Outcomes of patients with metastatic phaeochromocytoma and paraganglioma: a systematic review and meta-analysis. Clin. Endocrinol. 87(5), 440–450 (2017)
T.I. Korevaar, A.B. Grossman, Pheochromocytomas and paragangliomas: assessment of malignant potential. Endocrine 40(3), 354–365 (2011)
E. Patterson et al. The microRNA expression changes associated with malignancy and SDHB mutation in pheochromocytoma. Endocr.-Relat. Cancer 19(2), 157–166 (2012)
S. Azarbarzin et al. The value of MiR-383, an intronic MiRNA, as a diagnostic and prognostic biomarker in intestinal-type gastric cancer. Biochem. Genet. 55(3), 244–252 (2017)
C. Zhu, Q. Huang, H. Zhu, miR-383 inhibited the cell cycle progression of gastric cancer cells via targeting cyclin E2. DNA Cell. Biol. 38(8), 849–856 (2019)
T. Wang et al. CAMK2N1 inhibits prostate cancer progression through androgen receptor-dependent signaling. Oncotarget 5(21), 10293–10306 (2014)
F.H. Khan et al. RD3 loss dictates high-risk aggressive neuroblastoma and poor clinical outcomes. Oncotarget 6(34), 36522–36534 (2015)
Funding
This research was supported by the Intramural Research Program of the NIH, Eunice Kennedy Shriver National Institute of Child Health and Human Development (NICHD).
Author information
Authors and Affiliations
Contributions
Conception and design: S.G., K.P. Development of methodology: S.G., B.Z. Acquisition of data and experimentation (acquired and managed patients, performed qRT-PCR, etc.): B.Z., T.-T.H., K.P. Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): S.G., B.Z. Writing and review of the manuscript: S.G., B.Z., L.M., A.J., S.T., M.K., M.P., T.P., T.-T.H., S.D., M.A.Z., N.N., U.T.S., D.T., K.P. Study supervision: K.P.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Ghosal, S., Zhu, B., Huynh, TT. et al. A long noncoding RNA–microRNA expression signature predicts metastatic signature in pheochromocytomas and paragangliomas. Endocrine 75, 244–253 (2022). https://doi.org/10.1007/s12020-021-02857-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12020-021-02857-0