Abstract
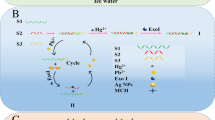
A novelty aptasensor for ultrasensitive detection of Hg2+ is developed, exploiting the combination of plasmonic properties of gold nanoparticles (AuNPs) and exonuclease III (Exo III)-assisted target recycling for signal amplification. In the presence of Hg2+, a DNA duplex can be formed due to the strong coordination of Hg2+ and T bases of single-stranded DNA (ssDNA) probe. Exo III digests the DNA duplex from the 3′ to 5′ direction, resulting in the releasing of Hg2+. Then, the released Hg2+ binds with another ssDNA probe through T-Hg2+-T coordination. After Exo III-assisted Hg2+ cycles, numerous ssDNA probes are exhausted, which promotes poly(diallyldimethylammonium chloride) (PDDA)-induced AuNP aggregation, leading to an obvious color change and aggregation-induced plasmon red shift of AuNPs (from 520 to 610 nm). Therefore, this biosensor is ultrasensitive, which is applicable to the detection of trace level of Hg2+ with a linear range from 5 pM to 0.6 nM and an ultralow detection limit of 0.2 pM. Furthermore, it enables visual detection of Hg2+ as low as 50 pM by the naked eye. More importantly, the assay can be applied to the reliable determination of spiked Hg2+ in sea water samples with good recovery.





Similar content being viewed by others
References
Yao L, Gao S, Liu S, Bi Y, Zheng L. Single-atom enzyme functionalized solution-gated graphene transistor for real-time detection of mercury ion. ACS Appl Mater Interfaces. 2020;12(5):6268–75.
Harris HH, Pickering IJ, George GN. The chemical form of mercury in fish. Science. 2003;301(5637):1203.
Wu H, Tong C. Dual-emission fluorescent probe for the simultaneous detection of nitrite and mercury (II) in environmental water samples based on the Tb3+-modified carbon quantum dot/3-aminophenylboronic acid hybrid. Anal Chem. 2020;92(13):8859–66.
Rahman GM, Wolle MM, Fahrenholz T, Kingston HS, Pamuku M. Measurement of mercury species in whole blood using speciated isotope dilution methodology integrated with microwave-enhanced solubilization and spike equilibration, headspace–solid-phase microextraction, and GC-ICP-MS analysis. Anal Chem. 2014;86(12):6130–7.
Chen X, Han C, Cheng H, Wang Y, Liu J, Xu Z, et al. Rapid speciation analysis of mercury in seawater and marine fish by cation exchange chromatography hyphenated with inductively coupled plasma mass spectrometry. J Chromatogr A. 2013;1314:86–93.
Gao Z, Ma X. Speciation analysis of mercury in water samples using dispersive liquid-liquid microextraction combined with high-performance liquid chromatography. Anal Chim Acta. 2011;702(1):50–5.
Yuan Y, Wu Y, Wang H, Tong Y, Sheng X, Sun Y, et al. Simultaneous enrichment and determination of cadmium and mercury ions using magnetic PAMAM dendrimers as the adsorbents for magnetic solid phase extraction coupled with high performance liquid chromatography. J Hazard Mater. 2020;386:121658.
Butler OT, Cairns W, Cook JM, Davidson CM. Atomic spectrometry update. Environmental analysis. J Anal At Spectrom. 2012;26:187–221.
Martinis EM, Bertón P, Olsina RA, Altamirano JC, Wuilloud RG. Trace mercury determination in drinking and natural water samples by room temperature ionic liquid based-preconcentration and flow injection-cold vapor atomic absorption spectrometry. J Hazard Mater. 2009;167(1–3):475–81.
Song C, Zhang Y, Li X, Ouyang G, Cui J, Zhang L, et al. Morphology-maintaining synthesis of copper hydroxy phosphate@metal–organic framework composite for extraction and determination of trace mercury in rice. Food Chem. 2021;343:128508.
Ma Y, Geng F, Wang Y, Xu M, Shao C, Qu P, et al. Novel strategy to improve the sensing performances of split ATP aptamer based fluorescent indicator displacement assay through enhanced molecular recognition. Biosens Bioelectron. 2019;134:36–41.
Kolm C, Cervenka I, Aschl UJ, Baumann N, Jakwerth S, Krska R, et al. DNA aptamers against bacterial cells can be efficiently selected by a SELEX process using state-of-the art qPCR and ultra-deep sequencing. Sci Rep. 2020;10(1):20917.
Li H, Chang J, Gai P, Feng L. Label-free and ultrasensitive biomolecule detection based on aggregation induced emission fluorogen via target-triggered hemin/G-quadruplex-catalyzed oxidation reaction. ACS Appl Mater Interfaces. 2018;10(5):4561–8.
Wang X, Lv W, Wu J, Li H, Li F. In situ generated nanozyme-initiated cascade reaction for amplified surface plasmon resonance sensing. Chem Commun. 2020;56(33):4571–4.
Ono A, Togashi H. Highly selective oligonucleotide-based sensor for mercury(II) in aqueous solutions. Angew Chem Int Ed Eng. 2004;43:4300–2.
Miyake Y, Togashi H, Tashiro M, Yamaguchi H, Oda S, Kudo M, et al. MercuryII-mediated formation of thymine-HgII-thymine base pairs in DNA duplexes. J Am Chem Soc. 2006;128(7):2172–3.
Tanaka Y, Oda S, Yamaguchi H, Kondo Y, Kojima C, Ono A. 15N-15N J-coupling across HgII: direct observation of HgII-mediated T-T base pairs in a DNA duplex. J Am Chem Soc. 2007;129(2):244–5.
Wang S, Lin B, Chen L, Li N, Xu J, Wang J, et al. Branch-migration based fluorescent probe for highly sensitive detection of mercury. Anal Chem. 2018;90(20):11764–9.
Li Z, Sun H, Ma X, Su R, Sun R, Yang C, et al. Label-free fluorescence “turn-on” strategy for mercury (II) detection based on the T-Hg2+-T configuration and the DNA-sensitized luminescence of terbium (III). Anal Chim Acta. 2020;1099:136–44.
Kong RM, Ma L, Han X, Ma C, Qu F, Xia L. Hg2+-mediated stabilization of G-triplex based molecular beacon for label-free fluorescence detection of Hg2+, reduced glutathione, and glutathione reductase activity. Spectrochim Acta A Mol Biomol Spectrosc. 2020;228:117855.
Hu PP, Liu N, Wu KY, Zhai LY, Xie BP, Sun B, et al. Successive and specific detection of Hg2+ and I– by a DNA@MOF biosensor: experimental and simulation studies. Inorg Chem. 2018;57(14):8382–9.
Chen J, Pan J, Shu C. A naked-eye colorimetric sensor for Hg2+ monitoring with cascade signal amplification based on target-induced conjunction of split DNAzyme fragments. Chem Commun. 2017;53(73):10224–7.
Li X, Du Z, Lin S, Tian J, Tian H, Xu W. Exo III and TdT dependent isothermal amplification (ETDA) colorimetric biosensor for ultra-sensitive detection of Hg2+. Food Chem. 2020;316:126980.
Memon AG, Zhou X, Liu J, Wang R, Liu L, Yu B, et al. Utilization of unmodified gold nanoparticles for label-free detection of mercury (II): insight into rational design of mercury-specific oligonucleotides. J Hazard Mater. 2017;321:417–23.
Xing Y, Zhu Q, Zhou X, Qi P. A gold nanoparticle-based colorimetric mercury(II) biosensor using a DNA probe with phosphorothioate RNA modification and exonuclease III-assisted signal amplification. Microchim Acta. 2020;187(4):1–8.
Bala A, Górski Ł. Peptide nucleic acid as a selective recognition element for electrochemical determination of Hg2+. Bioelectrochemistry. 2017;119:189–95.
Xiong E, Wu L, Zhou J, Yu P, Zhang X, Chen J. A ratiometric electrochemical biosensor for sensitive detection of Hg2+ based on thymine-Hg2+-thymine structure. Anal Chim Acta. 2015;853:242–8.
Ma N, Ren X, Wang H, Kuang X, Fan D, Wu D, et al. Ultrasensitive controlled release aptasensor using thymine-Hg2+-thymine mismatch as a molecular switch for Hg2+ detection. Anal Chem. 2020;92(20):14069–75.
Zhang L, Chang H, Hirata A, Wu H, Xue QK, Chen M. Nanoporous gold based optical sensor for sub-ppt detection of mercury ions. ACS Nano. 2013;7(5):4595–600.
Liu CW, Hsieh YT, Huang CC, Lin ZH, Chang HT. Detection of mercury(II) based on Hg2+-DNA complexes inducing the aggregation of gold nanoparticles. Chem Commun. 2008;19:11764–9.
Memon AG, Xing Y, Zhou X, Wang R, Liu L, Zeng S, et al. Ultrasensitive colorimetric aptasensor for Hg2+ detection using Exo-III assisted target recycling amplification and unmodified AuNPs as indicators. J Hazard Mater. 2020;384:120948.
Song X, Wang Y, Liu S, Zhang X, Wang H, Wang J, et al. Colorimetric and visual mercury(II) assay based on target-induced cyclic enzymatic amplification, thymine-Hg(II)-thymine interaction, and aggregation of gold nanoparticles. Microchim Acta. 2019;186(2):1–7.
Zhu Y, Cai Y, Zhu Y, Zheng L, Ding J, Quan Y, et al. Highly sensitive colorimetric sensor for Hg2+ detection based on cationic polymer/DNA interaction. Biosens Bioelectron. 2015;69:174–8.
Gu C, Kong X, Liu X, Gai P, Feng L. Enzymatic biofuel-cell-based self-powered biosensor integrated with DNA amplification strategy for ultrasensitive detection of single-nucleotide polymorphism. Anal Chem. 2019;91(13):8697–704.
Yu L, Wei L, Hui X, Hou C, Bai L, Wang W. Label-free detection of Hg2+ based on Hg2+-triggered toehold binding, exonuclease III assisted target recycling and hybridization chain reaction. Sensor Actuat B-Chem. 2017;248:411–8.
Gan X, Zhao H, Chen S, Quan X. Electrochemical DNA sensor for specific detection of picomolar Hg(II) based on exonuclease III-assisted recycling signal amplification. Analyst. 2015;140(6):2029–36.
Liu S, Lin Y, Wang L, Liu T, Cheng C, Wei W, et al. Exonuclease III-aided autocatalytic DNA biosensing platform for immobilization-free and ultrasensitive electrochemical detection of nucleic acid and protein. Anal Chem. 2014;86(8):4008–15.
Zuo X, Xia F, Xiao Y, Plaxco KW. Sensitive and selective amplified fluorescence DNA detection based on exonuclease III-aided target recycling. J Am Chem Soc. 2010;132(6):1816–8.
Zhao Y, Gao XY, Wang H, Wang J, Zhou J, Zhao W, et al. Ultrasensitive detection of microRNA via a Au@Ag nanosnowman. Anal Chem. 2019;91(24):15988–92.
Feng Q, Qin L, Wang M, Wang P. Signal-on electrochemical detection of DNA methylation based on the target-induced conformational change of a DNA probe and exonuclease III-assisted target recycling. Biosens Bioelectron. 2020;149:111847.
Song X, Yu W, Su L, Xue Z, Wang H, Wang J, et al. Ultrasensitive electrochemical detection of Hg2+ based on an Hg2+-triggered exonuclease III-assisted target recycling strategy. Analyst. 2018;143(23):4008–15.
Sang F, Yin S, Pan J, Liu D, Zhang Z. Colorimetric determination of DNA using an aptamer and plasmonic nanoplatform. Microchim Acta. 2020;187(7):1–7.
Sang F, Zhang X, Liu J, Yin S, Zhang Z. A label-free hairpin aptamer probe for colorimetric detection of adenosine triphosphate based on the anti-aggregation of gold nanoparticles. Spectrochim Acta A Mol Biomol Spectrosc. 2019;217:122–7.
Sang F, Liu J, Zhang X, Pan J. An aptamer-based colorimetric Pt (II) assay based on the use of gold nanoparticles and a cationic polymer. Microchim Acta. 2018;185(5):1–8.
Zhang Y, Xiao JY, Zhu Y, Tian LJ, Wang WK, Zhu TT, et al. A fluorescence sensor based on biosynthetic CdSe/CdS quantum dots and liposome carrier signal amplification for mercury detection. Anal Chem. 2020;92(5):3990–7.
Funding
This work was funded by the Natural Science Foundation of China (NSFC) (No. 21407035), Shandong Provincial Natural Science Foundation (ZR2014BM021), Technology and Development Program of Weihai (2014DXGJ15), and HIT-NSRIF (2011101).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(DOCX 1291 kb)
Rights and permissions
About this article
Cite this article
Sang, F., Yin, S., Pan, J. et al. Ultrasensitive colorimetric strategy for Hg2+ detection based on T-Hg2+-T configuration and target recycling amplification. Anal Bioanal Chem 413, 7001–7007 (2021). https://doi.org/10.1007/s00216-021-03657-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-021-03657-1