Abstract
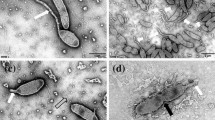
A novel actinobacterium NJES-13T was isolated from the gut of Antarctic emperor penguin Aptenodytes forsteri. The new isolate produces bioactive gephyromycin metabolites and exopolysaccharides (EPS). Cells were Gram-negative, motile with the peritrichous flagella, and with a faint layer of extracellular slime. Colonies were yellow when grown on marine agar, ISP1, 2, 4 and TSA media. The strain developed clusters of coccoid, and divided by binary fission in the early phase of growth. The cell clusters were gradually disrupted during the stationary phase and formed short rod-shape cells which were interconnected by viscous EPS showing a three-dimensional net-like morphology, and contained polyhydroxyalkanoates (PHA) granules inside the cells. Growth of strain NJES-13T was observed at 15–45 °C, at pH 6.0–9.0 with 0.5–9.0% (w/v) NaCl. The complete genomic size of strain NJES-13T was 3.45 Mb with a DNA G + C content of 67.0 mol%. The combined polyphasic taxonomic characterizations presented in this study unequivocally separated strain NJES-13T from all known genera in the family Dermatophilaceae. Thus, strain NJES-13T represents a novel species of a new genus, for which the name Gephyromycinifex aptenodytis gen. nov., and sp. nov. is proposed. The type strain is NJES-13T (= CCTCC 2019007T = KCTC 49281T). Genetic prediction of secondary metabolite biosynthesis revealed a 44.5 kb-long biosynthetic gene cluster (BGC) of type III polyketide synthase (PKS) as well as four other BGCs, indicating its great potential to produce novel bioactive metabolites derived from the gut microbiota of animals living in the extreme habitats in the Antarctica.




Similar content being viewed by others
Data availability
All data have been made fully available to the public.
Abbreviations
- AAI :
-
Average amino acid identity
- ANI :
-
Average nucleotide identity
- antiSMASH :
-
Antibiotics and Secondary Metabolite Analysis Shell
- BGCs :
-
Biosynthetic gene clusters
- dDDH :
-
Digital DNA-DNA hybridization
- DPG :
-
Diphosphatidylglycerol
- DSMZ :
-
Deutsche Sammlung von Mikro organismen und Zellkulturen
- eggNOG :
-
Evolutionary genealogy of genes: non-supervised orthologous groups
- EPS :
-
Exopolysaccharides
- GPM :
-
Gephyromycin
- GL :
-
Glycolipid
- GTDB-Tk :
-
Genome taxonomy database toolkit
- gyrB :
-
Gyrase B-encoding gene
- RP-HPLC :
-
Reversed-phase high performance liquid chromatography
- MA :
-
Marine agar
- MEGA :
-
Molecular evolutionary genetics analysis
- ML :
-
Maximum-likelihood
- MP :
-
Maximum-parsimony
- NJ :
-
Neighbour-joining
- PG :
-
Phosphatidylglycerol
- PHA :
-
Polyhydroxyalkanoates
- PI :
-
Phosphatidylinositol
- PL :
-
Phospholipids
- POCP :
-
Percentage of conserved proteins
- TLC :
-
Thin-layer chromatography
- UBCG :
-
Up-to-date bacterial core gene set
References
Abdelmohsen UR, Bayer K, Hentschel U (2014) Diversity, abundance and natural products of marine sponge-associated actinomycetes. Nat Prod Rep 31:381–399
Almeida A, Mitchell AL, Boland M, Forster SC, Gloor GB, Tarkowska A, Lawley TD, Finn RD (2019) A new genomic blueprint of the human gut microbiota. Nature 568:499–504
Azuma R, Ung-Bok B, Murakami S, Ishiwata H, Osaki M, Shimada N, Ito Y, Miyagawa E, Makino T, Kudo T, Takahashi Y, Yano I, Murata R, Yokoyama E (2013) Tonsilliphilus suis gen. nov., sp. nov., causing tonsil infections in pigs. Int J Syst Evol Microbiol 63:2545–2552
Baerson SR, Schröder J, Cook D, Rimando AM, Pan Z, Dayan FE, Noonan BP, Duke SO (2010) Alkylresorcinol biosynthesis in plants: new insights from an ancient enzyme family? Plant Signal Behav 5:1286–1289
Binda C, Lopetuso LR, Rizzatti G, Gibiino G, Cennamo V, Gasbarrini A (2018) Actinobacteria: a relevant minority for the maintenance of gut homeostasis. Dig Liver Dis 50:421–428
Blin K, Shaw S, Kloosterman AM, Charlop-Powers Z, van Wezel GP, Medema MH, Weber T (2021) AntiSMASH 6.0: improving cluster detection and comparison capabilities. Nucleic Acids Res 12:gkab335
Blunt JW, Copp BR, Keyzers RA, Munro MH, Prinsep MR (2013) Marine natural products. Nat Prod Rep 30:237–323
Chaumeil PA, Mussig AJ, Hugenholtz P, Parks DH (2019) GTDB-Tk: a toolkit to classify genomes with the genome taxonomy database. Bioinformatics 36:1925–1927
Chun J, Oren A, Ventosa A, Christensen H, Arahal DR, da Costa MS, Rooney AP, Yi H, Xu XW, De Meyer S, Trujillo ME (2018) Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol 68:461–466
Collins M (1985) Isoprenoid quinone analyses in bacterial classification and identification. In: Goodfellow M, Minnikin DE (eds) Chemical methods in bacterial systematics (society for applied bacteriology technical series no. 20). Academic Press, London, pp 267–287
Collins MD, Routh J, Saraswathy A, Lawson PA, Schumann P, Welinder-Olsson C, Falsen E (2004) Arsenicicoccus bolidensis gen. nov., sp. nov., a novel actinomycete isolated from contaminated lake sediment. Int J Syst Evol Microbiol 54:605–608
Dang PH, Nguyen LTT, Nguyen HTT, Le TH, Do TNV, Nguyen HX, Le ND, Nguyen MTT, Nguyen NT (2019) A new dimeric alkylresorcinol from the stem barks of Swintonia floribunda (Anacardiaceae). Nat Prod Res 33:2883–2889
Ding WJ, Ji YY, Jiang YJ, Ying WJ, Fang ZY, Gao TT (2020) Gephyromycin C, a novel small-molecule inhibitor of heat shock protein Hsp90, induces G2/M cell cycle arrest and apoptosis in PC3 cells in vitro. Biochem Biophys Res Commun 531:377–382
Duan Y, Jiang Z, Wu Z, Sheng Z, Yang X, Sun J, Zhang X, Yang Q, Yu X, Yan J (2020) Limnobacter alexandrii sp. nov., a thiosulfate-oxidizing, heterotrophic and EPS-bearing Burkholderiaceae isolated from cultivable phycosphere microbiota of toxic Alexandrium catenella LZT09. Antonie Van Leeuwenhoek 113:1689–1698
Gao HM, Xie PF, Zhang XL, Yang Q (2021) Isolation, phylogenetic and gephyromycin metabolites characterization of new exopolysaccharides-bearing Antarctic actinobacterium from faeces of emperor penguin. Mar Drugs 19:458
Gerhard B, Gerhard L, Kadja M, Andreas H, Tobias AMG, Anke D, Alan TB, James EMS, Niko K, Werner EGM (2005) Gephyromycin, the first bridged angucyclinone, from Streptomyces griseus strain NTK 14. Phytochem 66:1366–1373
Giovannoni SJ and Nemergut D (2014) Microbes ride the current. Science 345:1246
Gordon MA (1964) The genus Dermatophilus. J Bacteriol 88:509–522
Hamada M, Iino T, Iwami T, Harayama S, Tamura T, Suzuki K (2010) Mobilicoccus pelagius gen. nov., sp. nov. and Piscicoccus intestinalis gen. nov., sp. nov., two new members of the family Dermatophilaceae, and reclassification of Dermatophilus chelonae (Masters et al. 1995) as Austwickia chelonae gen. nov., comb. nov. J Gen Appl Microbiol 56:427–436
Huerta-Cepas J, Szklarczyk D, Heller D, Hernández-Plaza A, Forslund SK, Cook H, Mende DR, Letunic I, Rattei T, Jensen LJ, von Mering C, Bork P (2019) eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res 47:D309–D314
Kates M (1986) Lipid extraction procedures. In: Burdun RH, van Knippenberg PH (eds) Laboratory techniques in biochemistry and molecular biology. Elsevier, Amsterdam, pp 350–351
Kern DL, Schaumberg JP, Hokanson GC, French JC (1986) PD 116,779, a new antitumor antibiotic of the benz[a]anthraquinone class. J Antibiot 39:469–470
Landry ZC, Vergin K, Mannenbach C, Block S, Yang Q, Blainey P, Carlson C, Giovannoni S (2018) Optofluidic single-cell genome amplification of sub-micron bacteria in the ocean subsurface. Front Microbiol 9:1152
Li S, Yang B, Tan GY, Ouyang LM, Qiu S, Wang W, Xiang W, Zhang L (2021) Polyketide pesticides from actinomycetes. Curr Opin Biotechnol 69:299–307
Liu WT, Hanada S, Marsh TL, Kamagata Y, Nakamura K (2002) Kineosphaera gen. nov., sp. nov., a novel Gram-positive polyhydroxyalkanoate-accumulating coccus isolated from activated sludge. Int J Syst Evol Microbiol 52:1845–1849
Masters AM, Ellis TM, Carson JM, Sutherland SS, Gregory AR (1995) Dermatophilus chelonae sp. nov., isolated from chelonids in Australia. Int J Syst Bacteriol 45:50–56
Méndez C, Künzel E, Lipata F, Lombó F, Cotham W, Walla M, Bearden DW, Braña AF, Salas JA, Rohr J (2002) Oviedomycin, an unusual angucyclinone encoded by genes of the oleandomycin-producer Streptomyces antibioticus ATCC11891. J Nat Prod 65:779–782
Na SI, Kim YO, Yoon SH, Ha SM, Baek I, Chun J (2018) UBCG: up-to-date bacterial core gene set and pipeline for phylogenomic tree reconstruction. J Microbiol 56:281–285
Nouioui I, Carro L, García-López M, Meier-Kolthoff JP, Woyke T, Kyrpides NC, Pukall R, Klenk HP, Goodfellow M, Göker M (2018) Genome-based taxonomic classification of the phylum Actinobacteria. Front Microbiol 9:2007
Qin QL, Xie BB, Zhang XY, Chen XL, Zhou BC, Zhou J, Oren A, Zhang YZ (2014) A proposed genus boundary for the prokaryotes based on genomic insights. J Bacteriol 196:2210–2215
Richter M, Rosselló-Móra R (2009) Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA 106:19126–19131
Salam N, Jiao JY, Zhang XT, Li WJ (2020) Update on the classification of higher ranks in the phylum Actinobacteria. Int J Syst Evol Microbiol 70:1331–1355
Takeda K, Kang Y, Yazawa K, Gonoi T, Mikami Y (2010) Phylogenetic studies of Nocardia species based on gyrB gene analyses. J Med Microbiol 59:165–171
van Saceghem R (1915) Dermatose contagieuse. Bull Soc Pathol Exot 8:354–359
van Bergeijk DA, Terlouw BR, Medema MH, van Wezel GP (2020) Ecology and genomics of Actinobacteria: new concepts for natural product discovery. Nat Rev Microbiol 18:546–558
Wang Y, Coleman-Derr D, Chen G, Gu YQ (2015) Orthovenn: a web server for genome wide comparison and annotation of orthologous clusters across multiple species. Nucleic Acids Res 43:W78–W84
Wirth JS, Whitman WB (2018) Phylogenomic analyses of a clade within the roseobacter group suggest taxonomic reassignments of species of the genera Aestuariivita, Citreicella, Loktanella, Nautella, Pelagibaca, Ruegeria, Thalassobius, Thiobacimonas and Tropicibacter, and the proposal of six novel genera. Int J Syst Evol Microbiol 68:2393–2411
Yamamoto S, Harayama S (1995) PCR amplification and direct sequencing of gyrB genes with universal primers and their application to the detection and taxonomic analysis of Pseudomonas putida strains. Appl Environ Microbiol 61:1104–1109
Yang Q, Jiang ZW, Huang CH, Zhang RN, Li LZ, Yang G, Feng LJ, Yang GF, Zhang H, Zhang XL, Mu J (2018a) Hoeflea prorocentri sp. nov., isolated from a culture of the marine dinoflagellate Prorocentrum mexicanum PM01. Antonie Van Leeuwenhoek 111:1845–1853
Yang Q, Zhang XL, Li L, Zhang RN, Feng LJ, Mu J (2018b) Ponticoccus alexandrii sp. nov., a novel bacterium isolated from the marine toxigenic dinoflagellate Alexandrium minutum. Antonie Van Leeuwenhoek 111:995–1000
Yang Q, Jiang Z, Zhou X, Xie Z, Wang Y, Wang D, Feng L, Yang G, Ge Y, Zhang X (2020a) Saccharospirillum alexandrii sp. nov., isolated from the toxigenic marine dinoflagellate Alexandrium catenella LZT09. Int J Syst Evol Microbiol 70:820–826
Yang Q, Jiang Z, Zhou X, Zhang R, Wu Y, Lou L, Ma Z, Wang D, Ge Y, Zhang X, Yu X (2020b) Nioella ostreopsis sp. nov., isolated from toxic dinoflagellate, Ostreopsis lenticularis. Int J Syst Evol Microbiol 70:759–765
Yang Q, Jiang Z, Zhou X, Zhang R, Xie Z, Zhang S, Wu Y, Ge Y, Zhang X (2020c) Haliea alexandrii sp. nov., isolated from phycosphere microbiota of the toxin-producing dinoflagellate Alexandrium catenella. Int J Syst Evol Microbiol 70:1133–1138
Yang X, Jiang ZW, Zhang J, Zhou X, Zhang XL, Wang L, Yu T, Wang Z, Bei J, Dong B (2020d) Mesorhizobium alexandrii sp. nov., isolated from phycosphere microbiota of PSTs-producing marine dinoflagellate Alexandrium minutum amtk4. Antonie Van Leeuwenhoek 113:907–917
Yang Q, Feng Q, Zhang BP, Gao JJ, Sheng Z, Xue QP, Zhang XL (2021a) Marinobacter alexandrii sp. nov., a novel yellow-pigmented and algae growth-promoting bacterium isolated from marine phycosphere microbiota. Antonie Van Leeuwenhoek 114:709–718
Yang Q, Ge YM, Iqbal NM, Yang X, Zhang XL (2021b) Sulfitobacter alexandrii sp. nov., a new microalgae growth-promoting bacterium with exopolysaccharides bioflocculanting potential isolated from marine phycosphere. Antonie Van Leeuwenhoek 114:1091–1106
Yarza P, Ludwig W, Euzéby J, Amann R, Schleifer KH, Glöckner FO, Rosselló-Móra R (2010) Update of the All-species living tree project based on 16S and 23S rRNA sequence analyses. Syst Appl Microbiol 33:291–299
Zhang XL, Li GX, Ge YM, Iqbal MN, Yang X, Cui ZD, Yang Q (2021a) Sphingopyxis microcytisis sp. nov., a novel bioactive exopolysaccharides-bearing Sphingomonadaceae isolated from the Microcytis phycosphere. Antonie Van Leeuwenhoek 114:845–857
Zhang XL, Qi M, Li QH, Cui ZD, Yang Q (2021b) Maricaulis alexandrii sp. nov., a novel active bioflocculants-bearing and dimorphic prosthecate bacterium isolated from marine phycosphere. Antonie Van Leeuwenhoek 78:1648–1655
Zhou X, Zhang XA, Jiang ZW, Yang X, Zhang XL, Yang Q (2021) Combined characterization of a new member of Marivita cryptomonadis, strain LZ-15-2 isolated from cultivable phycosphere microbiota of toxic HAB dinoflagellate Alexandrium catenella LZT09. Braz J Microbiol 52:739–748
Acknowledgements
The authors sincerely acknowledge Prof. Aharon Oren at The Hebrew University of Jerusalem for his helping to name the new actinobacterium. We also thank Prof. Tingting Huang at the Laboratory of Molecular Microbiology, Shanghai Jiao Tong University for her kindly assistance for the gene cluster analysis.
Funding
This work was supported by the Fundamental Research Funds for Zhejiang Provincial Universities and Research Institutes (Grant No. 2021J020), the National Natural Science Foundation of China (Grant No. 41876114), and the Scientific Research Startup Foundation of Zhejiang Ocean University (Grant No. 11104150319/001).
Author information
Authors and Affiliations
Contributions
XLZ and QY conceived the project, WZZ, YMG, and HMG performed the experiments, WZZ, YMG and JD analyzed the data, and WZZ, XLZ and QY drafted the manuscript. All authors have read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
All the authors have declared no conflict of interest.
Consent to participate
All authors gave their consent to participate in this study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhu, WZ., Ge, YM., Gao, HM. et al. Gephyromycinifex aptenodytis gen. nov., sp. nov., isolated from gut of Antarctic emperor penguin Aptenodytes forsteri. Antonie van Leeuwenhoek 114, 2003–2017 (2021). https://doi.org/10.1007/s10482-021-01657-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-021-01657-w