- 1Mycopathology and Microbial Technology Laboratory, Centre of Advanced Study in Botany, Institute of Science, Banaras Hindu University, Varanasi, India
- 2Agricultural Research Organization, Rishon LeZion, Israel
- 3Center for Safe and Improved Food, Biorefining and Advanced Materials Research Center, Scotland’s Rural College, Edinburgh, United Kingdom
The endophytic fungus Diaporthe longicolla was isolated from the stem of Saraca asoca (Roxb.) Willd., commonly known as Ashok plant in India and Sri Lanka. Since no reports are available regarding epigenetic modulations by BRD4770 in microbial entities, D. longicolla was treated with different concentrations of BRD4770 for this purpose and evaluated for its antioxidant and antibacterial potential against five human pathogenic bacteria, Staphylococcus aureus, methicillin-resistant Staphylococcus aureus (MRSA), Shigella boydii, Klebsiella pneumoniae, and Escherichia coli. The crude extract obtained from cultures treated with 100 nM concentration of BRD4770 showed increased antioxidant activity and inhibition zone against S. aureus and MRSA, compared to the non-treated control. The composition of the non-treated and treated crude extract was analyzed, and induced compounds were identified with the help of Gas chromatography–mass spectrometry (GC-MS) and LC-ESI-MS/MS. LC-ESI-MS/MS analysis showed that berberine (antibacterial)-, caffeine-, and theobromine (antioxidant)-like compounds were induced in the BRD4770-treated crude extract. The presence of particular absorbance at a wavelength of 346.5 nm for berberine, 259.4 nm for caffeine, and 278.4 nm for theobromine in the reverse-phase high-performance liquid chromatography (HPLC) analysis of both BRD4770-treated crude metabolites and standard solution of the above compounds strongly supported the increased antibacterial and antioxidant activities that may be due to inducing the alterations in bioactivities of the BRD4770-treated culture.
Highlights
- An endophytic fungus (L3), isolated from the leaf tissues of Saraca asoca was identified as Diaporthe longicolla.
- The crude metabolite extracts of D. longicolla showed strong antioxidant and antibacterial properties.
- D. longicolla was treated with different concentrations of BRD4770 for evaluation of its antibacterial and antioxidant potential.
- The crude extract obtained from cultures treated with 100 nM concentration of BRD4770, showed increased inhibition zone against S. aureus and methicillin-resistant Staphylococcus aureus (MRSA), compared to the non-treated control.
- Antioxidant activity was also founded to increase in treated with 100 nM BRD4770 compared to the non-treated culture.
- The composition of non-treated as well as treated crude extract was analyzed and induced compounds were identified with the help of GC-MS and LC-ESI-MS/MS.
- LC-ESI-MS/MS analysis showed berberine (antibacterial), caffeine, and theobromine (antioxidant) like compounds, etc., were induced in BRD4770 treated crude extract.
- Presence of particular absorbance at wavelength 346.5 for berberine, 259.4 for caffeine and 278.4 nm for theobromine in the reverse phase HPLC analysis of both BRD4770 treated crude metabolites and standard solution confirms its presence.
Introduction
German scientist de Bary (1866) gave the term “endophytes” for any organism (bacteria, fungi, actinomycetes, etc.) that resides in healthy plant tissues internally, without causing any apparent disease symptoms to the host. Endophytes as an alternative source of plant-derived drugs like camptothecin, vincristine, vinblastine, rohitukine, azadirachtin, and piperine will reduce not only the price and crisis of host mimetic compounds but also the overexploitation of host plants (Kharwar et al., 2008, 2011, 2020; Verma et al., 2009, 2014; Singh et al., 2021). Diaporthe longicolla, a member of Ascomycota, belongs to class Dothideomycetes, order Diaporthales, and family Diaporthaceae (Bubák and Kabát, 1905). Various numbers of structurally diverse polyketides have also been reported from D. longicolla. These include cytoskyrins and mycotoxins that lead to DNA damage and inhibit the growth in Escherichia coli (Agusta et al., 2006). D. longicolla isolated from Annona squamosa produces compounds like diaporthemins A and B and flavomannin-6,6’-dimethylether that are antibacterial against Streptococcus pneumoniae (Ola et al., 2014). It has always been an interest of scientists to maximize the numbers and yield of compounds and also to receive cryptic metabolites from microbes by adopting different approaches such as substrate selection, co-culture, one strain many compounds (OSMAC), and epigenetic modulations.
In fungi, the biosynthetic potential of genes for secondary metabolites is typically organized in gene clusters, of which several clusters recognized for the biosynthesis of secondary metabolites remain silent (Rutledge and Challis, 2015). These silent or cryptic genes can be induced through epigenetic modulations to discover novel compounds. Epigenetic modulations can be used as a tool to stimulate an endophyte to produce more compounds with far greater bioactivities (Sharma et al., 2017). Treatment of fungi with histone deacetylase (HDAC) inhibitors like suberoylanilide hydroxamic acid (SAHA), sodium butyrate, and valproic acid, and DNA methyltransferase (DNMT) inhibitors like 5-aza-20-deoxycytidine 5-azacytidine, procainamide, procaine, and hydralazine has been shown to facilitate the induction of cryptic gene or activation of biosynthetic pathways of secondary metabolites. Similarly, methyl ester BRD4770 can significantly reduce cellular levels of H3K9me2 and H3K9me3, but may increase the cellular levels of H3K9me1 and activation of various cellular responses. These bioactive chemical probes alter the DNA methylation and histone modifications requisite for gene activation in the same way as the other chemical epigenetic modifiers (Meeran et al., 2010). Epigenetic influences of many chemical modulators have been much studied for the induction and activation of many known and new metabolites, but the effect of BRD4770 on the induction of secondary metabolites in fungi is still not known well. The BRD4770 is a novel histone methyltransferase inhibitor that can be instrumental in the epigenetic manipulation of silent gene clusters in fungal endophytes. This study aimed to use a histone methyltransferase (G9a), a specific probe BRD4770 either to enhance the activity of compounds or to isolate the cryptic metabolites from an endophytic fungus D. longicolla for the first time. The plant selected to isolate D. longicolla was Saraca asoca (Roxb.) Willd., a legendary and sacred tree of India, having one of the most fascinating flowers in the Indian range of flower essences.
Materials and Methods
Isolation, Identification, and Molecular Characterization of Endophytic Fungus
Isolation and identification of endophytic fungus D. longicolla were done from the mature healthy leaves of the S. asoca plant growing in the Botanical Garden of BHU (Banaras Hindu University) campus, Varanasi, India. Mature, green, asymptomatic leaves were randomly collected from the lowermost branches of the plant sealed in sterile polybags and kept in an icebox (4°C) until further processing. The modified protocol of Arnold and Lutzoni (2007) was adopted for surface sterilization (Petrini et al., 1992). Briefly, leaf was cut into 5 mm × 5 mm long segments and consecutively dipped in 70% ethanol for 1 min followed by 30 s in 4% sodium hypochlorite and finally rinsed thrice with autoclaved distilled water to remove excess surface sterilant. Under aseptic conditions, the tissue segments were dried before plating; four leaf segments were placed evenly onto PDA plates supplemented with streptomycin (50 μg/ml) to inhibit the growth of bacterial endophytes. Plates were sealed with parafilm and incubated in BOD incubator at 28°C ± 2°C for 5–7 days. Emerging fungal colonies were aseptically transferred to fresh PDA plates to obtain pure cultures. Primary identification of isolated endophytic fungus was done through morphology and reproductive structures observed under the microscope. Genotypic identification was done through total genomic DNA extraction (DNA kit, GeNei) from fresh mycelium cultured in potato dextrose broth (PDB). Polymerase chain reaction (PCR) was performed using extracted DNA with universal primers ITS1: TCCGT AGGTGAACCTGCGG and ITS4: TCCTCCGCTTGATATGC. Total PCR mixture for 25 μl, containing 1 μl of each primer, 0.33 μl (3 units/μl) of Taq polymerase, 0.5 μl of dNTPs, 2.5 μl of 10 × PCR buffer with 25 mM MgCl2, 1 μl (100 ng/ml) of DNA template, and 18.67 μl of milli-Q water. PCR was performed using MyCycler (Bio-Rad, Hercules, CA, United States), with pre-denaturation at 94°C for 4 min; 35 cycles of each denaturation was at 94°C for 1 min, and annealing was at 55°C for 1 min while extension was at 72°C for 1 min; final extension was at 72°C for 5 min. Amplified products were resolved by 1.5% (w/v) agarose using electrophoresis and gel was stained with ethidium bromide (0.5 mg/ml) for visual examination. PCR amplified DNA was further purified through gel excision method from Real Biotech Corporation (RBC, India) using a HiYield PCR DNA mini kit and sequenced by EDZ Daignon Pvt., Ltd., India. The obtained ITS rRNA gene sequences were used to retrieve similar sequences using the NCBI nBLAST program from the NCBI GenBank sequence database. The rRNA gene sequence was submitted to the NCBI GenBank database for accession number.
Preparation of BRD4770 Solution for Epigenetic Treatment
Stock solution (100 μM) was prepared by dissolving 4.13 mg of BRD4770 in 100 ml DMSO and filtered with 0.20 mm filter paper for removal of any contaminant. PDB medium (100 ml) was amended with BRD4770 separately in a series of final concentrations of 1, 10, 25, 50, 100, 200, 400, and 1,000 nM, respectively. Finally, each test culture was inoculated with the endophytic fungus and incubated at 26°C ± 2°C for 21 days. The treatments were done in triplicate for reproducibility.
Isolation of Secondary Metabolites
The selected culture of endophytic fungus was cultured in a 250-ml Erlenmeyer flask containing 100 ml of PDB medium for 21 days at 28°C and 150 rpm. The fermentation broth culture was filtered under clean and aseptic conditions. Extraction of the secondary metabolite from the filtrate was carried out thrice with the same volume of ethyl acetate as organic solvent. The solvent was added to the filtrate with 1:1 (v/v) ratio in a separating funnel, mixed properly for 10–15 min and left for 5–6 min until the formation of two clear immiscible layers. The metabolites containing an upper layer of solvent was collected and evaporated using a rotator vacuum evaporator (IKA, Germany) to yield the dried crude metabolites. The secondary metabolites obtained from different treatments were measured and dissolved in methanol to the final concentration of 10 mg/ml.
Chemicals and Materials
BRD4770 (Methyl-2-(benzoylamino)-1-(3-phenylpropyl)-1H-benzimidazole-5-carboxylate) (>98%) for the epigenetic modulation, caffeine (>98%), theobromine (>98%), and berberine (>98%) standards were purchased from Sigma-Aldrich Co. (St. Louis, MO, United States) for comparative analysis. Acetonitrile, methanol, 0.1% trifluoroacetic acid (TFA), and water (HPLC grade) were purchased from Merck Pvt., Ltd. (Mumbai, India). All the chemicals including solvents were of analytical grade.
Preparation of Reagents and Standards
For the mobile phase, 60 parts of 0.1% TFA and 40 parts of acetonitrile were mixed to get 0.1% TFA:acetonitrile (60:40 v/v) as a final solution. The mixed mobile phase was then filtered through nylon membrane vacuum filtration (0.22 μm) and degassed by sonication. Samples were analyzed on a detector at wavelength of 346.5 nm for berberine, 259.4 nm for caffeine, and 264.1 nm for theobromine and an injection volume of 10 μl using RP-C18 column (Waters, 4.6 mm × 100 mm, 3.5 μm) with a run time of 20 min.
Preparation of Standard Solutions
A standard stock solution was prepared by dissolving 100 mg of berberine, caffeine, and theobromine in a 100-ml volumetric flask containing 60 ml mobile phase and then sonicated for about 10 min and made up to 100 ml with mobile phase to get the primary standard stock solution containing 1 mg/ml of berberine, caffeine, and theobromine. Working standard solutions were prepared by further dilution with the mobile phase.
Detection of Berberine, Caffeine, and Theobromine in the Extract of D. longicolla by RP-HPLC
High-performance liquid chromatography analysis was performed for the treated crude extract and the non-treated crude extract of D. longicolla, using RP-C18 column (Waters, 4.6 mm × 100 mm, 3.5 μm, made in Netherlands). Pre-run sample stabilization was done for about 20 min with mobile phase consisting of 0.1% TFA:acetonitrile (60:40 v/v). The flow rate was maintained at 1.0 ml/min and the injected volume was 20 μl. The peak detection was done at 346.5 nm for berberine, 259.4 nm for caffeine, and 264.1 nm for theobromine, respectively, and the retention time (RT) was recorded duly maintaining the ambient experimental conditions. The presence of caffeine, theobromine, and berberine chromatogram in the BRD4770-treated sample was matched with the chromatogram of standards solution. The identification of each compound was based on a combination of RT, absorption maxima at a given wavelength, and chromatogram matching.
Antibacterial Activity
The metabolites extracted from all the different treatments were screened for the antibacterial activity against five human bacterial pathogens, methicillin-resistant Staphylococcus aureus (MRSA) IMS/GN11, Shigella boydii IMS/GN17, S. aureus ATCC 25923, Klebsiella pneumoniae, and E. coli IMS/GN19 using the disk diffusion method. The crude extract was loaded aseptically 100 μl (1 mg) to the sterilized filter paper disks and air-dried. A lawn of test bacterial culture was spread with a cotton swab onto the surface of solidified Mueller-Hinton agar (MHA) Petri plates. Sterile paper disks loaded with 1 mg of crude extract were placed over the lawn of bacterial culture on MHA plates. Positive control included similar paper disks impregnated with 10 μl of methanol and dried. Plates were incubated for 24 h at 35°C ± 2°C and then antibacterial activity including the minimum zone of inhibition was observed. Each test was performed in triplicate and the mean diameter of the minimum zones of inhibition with standard deviation was presented in the results (Table 1).
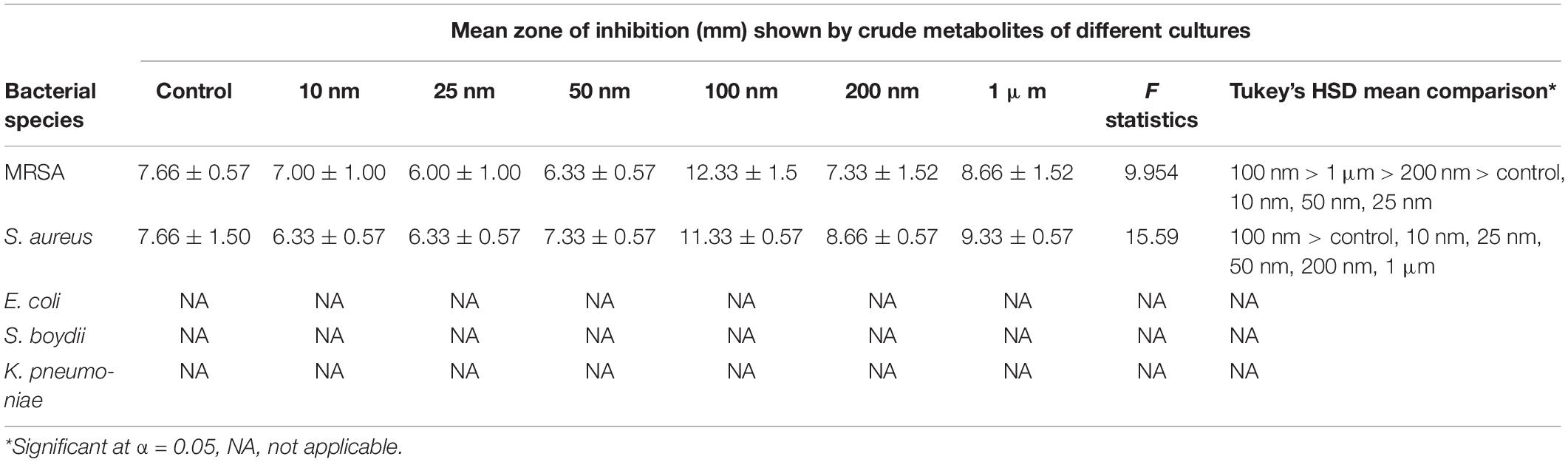
Table 1. Results of one-way ANOVA and Tukey’s HSD mean comparison examining the zone of inhibition against different bacteria using metabolite extracted from treated and non-treated fungus D. longicolla.
Antioxidant Activity
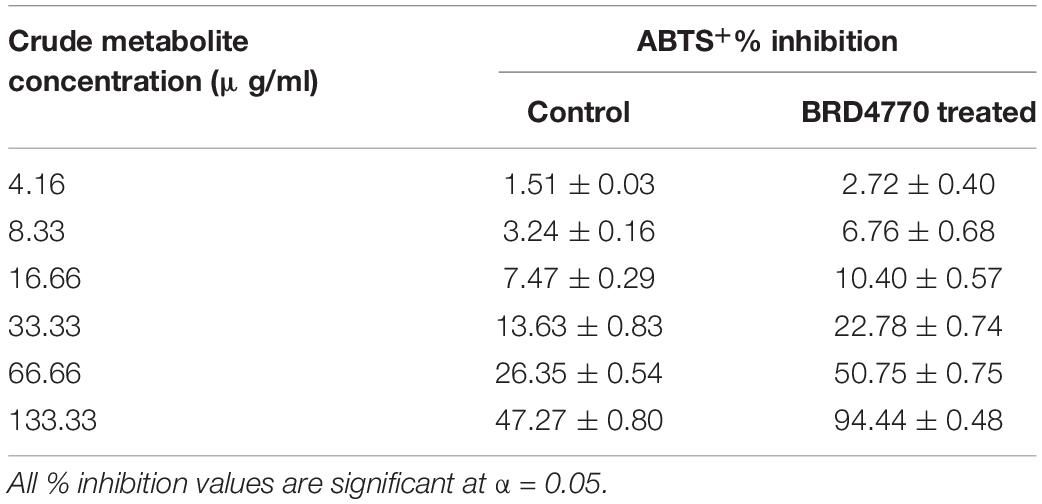
The antioxidant activity of treated as well as non-treated crude extract was determined using ABTS+ [2,2′-azino-bis(3-ethylbenzothiazoline-6-sulfonic acid)] radical scavenging method (Miller and Rice-Evans, 1997). Three milliliters of ABTS+ was allowed to react with various concentrations (12.5, 25, 50, 100, 200, and 400 μg/3 ml or 4.16, 8.33, 16.66, 33.33, 66.66, and 133.33/ml) (Figures 2A,B and Table 2) of treated and non-treated crude extract for 30 min, and absorbance was recorded at 734 nm. The O.D. of pure ABTS+ was used as control and percent inhibition was calculated using the following formula:
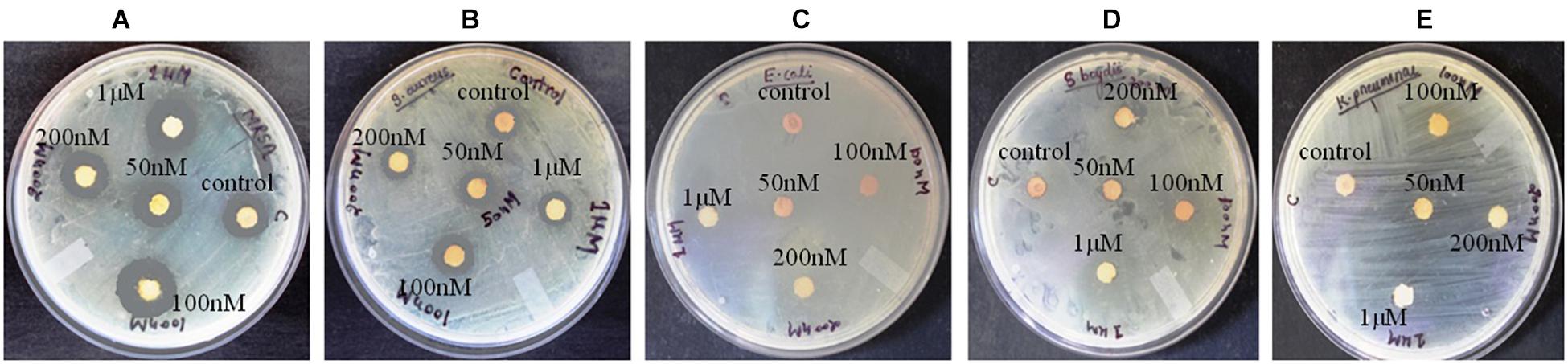
Figure 1. Antibacterial activity of crude compounds isolated from various concentration of BRD4770-treated and control (non-treated) cultures of D. longicolla. (A) Methicillin-resistant Staphylococcus aureus. (B) Staphylococcus aureus. (C) Escherichia coli. (D) Shigella boydii. (E) Klebsiella pneumoniae.

Figure 2. The ABTS+ radical scavenging method-based antioxidant activity of crude compounds in increasing concentration (μg/ml) isolated from D. longicolla (A) non-treated control and (B) BRD4770-treated cultures.
Where AControl and ASample or Reference are the absorbance of the ABTS+ in the control solution and sample or reference solution, respectively. The antioxidant capacities were calculated based on calibration curves plotted using different concentrations of the sample and reference solution. All tests were performed in triplicate.
Gas Chromatography–Mass Spectrometry Analysis Condition
Gas chromatography–mass spectrometry (GC-MS) analysis of the methanolic crude extracts was carried out in a (30 m 1D-0.25 mm film thickness-0.25 μm) capillary column of TSQ Duo Thermo Scientific a TG-5MS in BHU, Varanasi, India. The initial temperature of the instrument was set to 70°C and maintained for 3 min at this temperature. At the rate of an increase of 8°C/min, oven temperature was raised to 300°C at the end of this period, and maintained for 1 min. Helium flow rate was at 1 ml/min, and port of injection temperature was ensured at 250°C. The voltage of ionization was 70 eV. The samples were injected in a 40:1 split mode. Mass spectral scan range was set at 40–450 (m/z). The identification of bioactive compounds present in the crude extracts was performed by comparing the mass spectra with data from NIST05 (National Institute of Standards and Technology, United States) libraries. The name, molecular weight, and structure of the components of the test material were ascertained.
HPLC-ESI-MS/MS Analysis Condition
The HPLC-ESI-MS/MS of the extracted crude was done on a MS system using a high-performance liquid chromatography (HPLC) system (Waters model no. ACQ-TQD#QBB 1152) fitted with a C18 column having dimensions of 100 × 2.1, 1.7 μm. The mobile phase flow ramp rate was 0.45 ml/min and the injection volume of the sample was 10 μl. The mobile phase was acetonitrile/water (5:95, v/v), 5 mM ammonium acetate of pH 6.5 buffer, and methanol solutions. Run time was 30 min, resolution 1.2 nm, range 210–800 nm. MS scan was started at 150 mass and ended at 2000 mass under + ESI source and MS-MS collision energy was 2.0. HPLC eluted peaks were recorded at different RTs. The fractions were finally characterized by mass spectrometry. This analysis was carried out at Sophisticated Analytical Instrumentation Facility (SAIF), CSIR-Central Drug Research Institute (CDRI), Lucknow, India. The identification of bioactive compounds was performed by comparing the mass spectra obtained at a particular RT with mass spectra of standard compounds stored in the spectral library of mzCloud spectral Database.
Results
Based on microscopic as well as gene sequencing of ITS rRNA data, the isolate L3 was identified as D. longicolla with GenBank accession number MN480529.
Antibacterial Activity of Treated and Non-treated Culture
Antibacterial activity of crude extract obtained from the 100 nM BRD4770-treated culture showed significantly higher inhibition zone (11.33 ± 0.57 and 12.33 ± 1.5) against S. aureus and MRSA, respectively, as compared to the non-treated crude extract (Figure 1 and Table 1). No zone of inhibition was observed against the rest of the other bacterial pathogens used for the assay, neither in control nor in the treated ones (Table 1).
Antioxidant Activity of Crude Metabolite
ABTS+ radical scavenging activity was observed as% inhibition in crude metabolites of the control and treated culture (Figure 2 and Table 2). The calculated EC50 for the control was >133.33 μg/ml and that for the 100 nM BRD4770-treated culture was 66.66 μg/ml.
Characterization of Bioactive Compounds
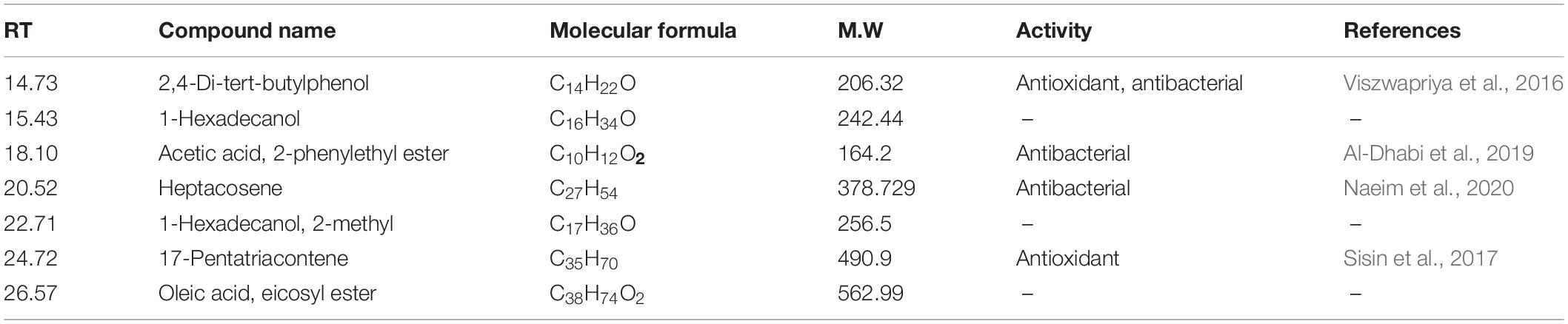
The GC-MS of the crude ethyl acetate extracts of the 100 nM BRD4770-treated cultures was found to induce in secondary metabolites biosynthesis, viz., benzoic acid, 4-methyl-2-trimethylsilyloxy, and (4-chloro-3-nitrophenyl)methanol, dimethylpentafluorophenylsilyl ether (Figure 3B), while crude secondary metabolites from the rest of the treatments including non-treated ones were approximately similar (Supplementary Figure 2) in composition (Figure 3A). Details of the compounds identified by GC-MS analysis in the (100 nM) treated and non-treated culture are provided in Tables 3, 4.
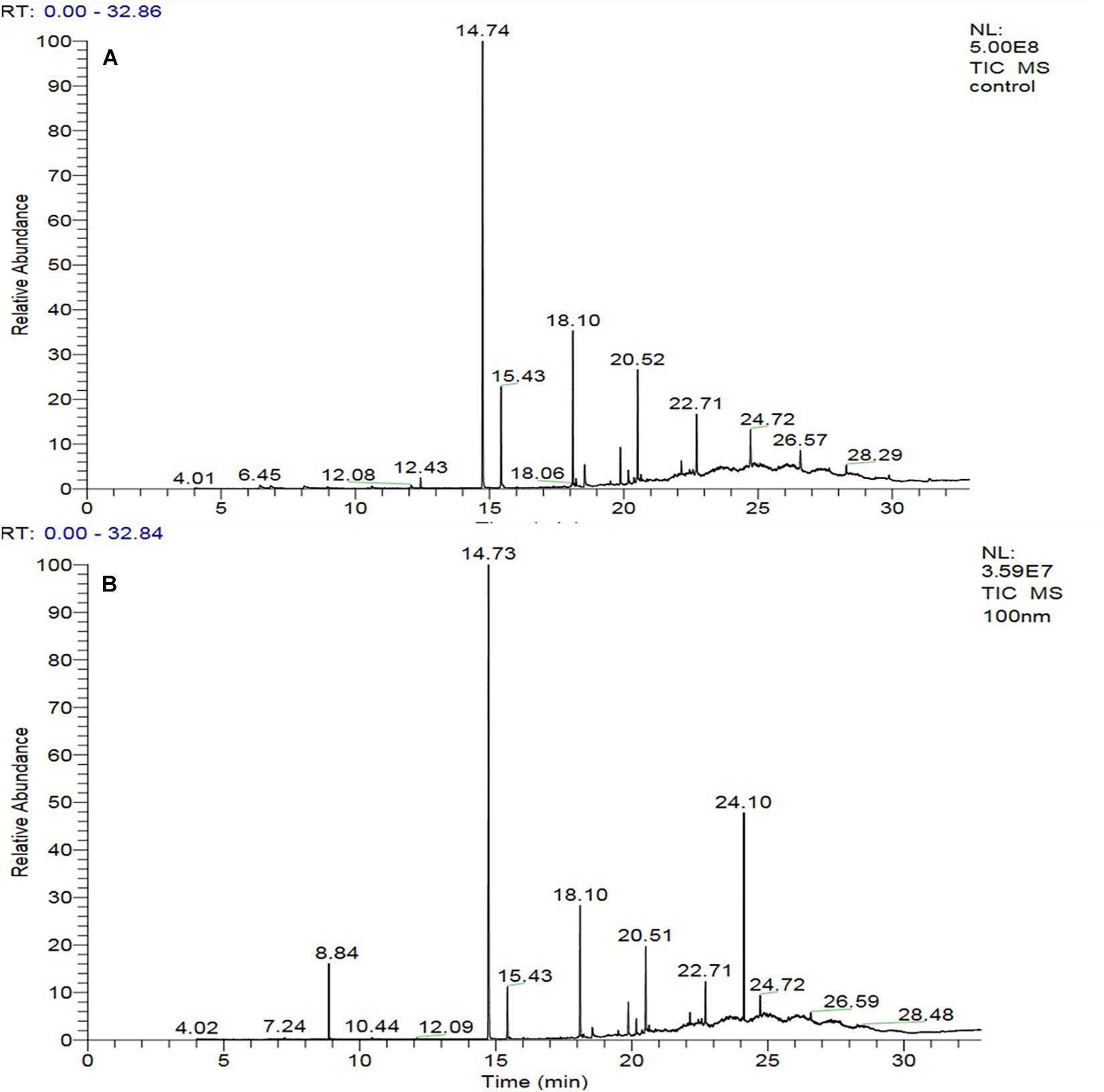
Figure 3. GC-MS chromatogram of crude metabolites of D. longicolla cultures (A) non-treated and (B) 100 nM BRD4770 treated.
Identification of Compounds by HPLC-ESI-MS/MS
In full scan mass spectra of fungal metabolite, [M-H]– was observed in negative ion ESI mode and [M+H]+ was observed in the positive ion ESI mode. The information on [M-H]– and [M+H]+ was used to determine molecular weight.
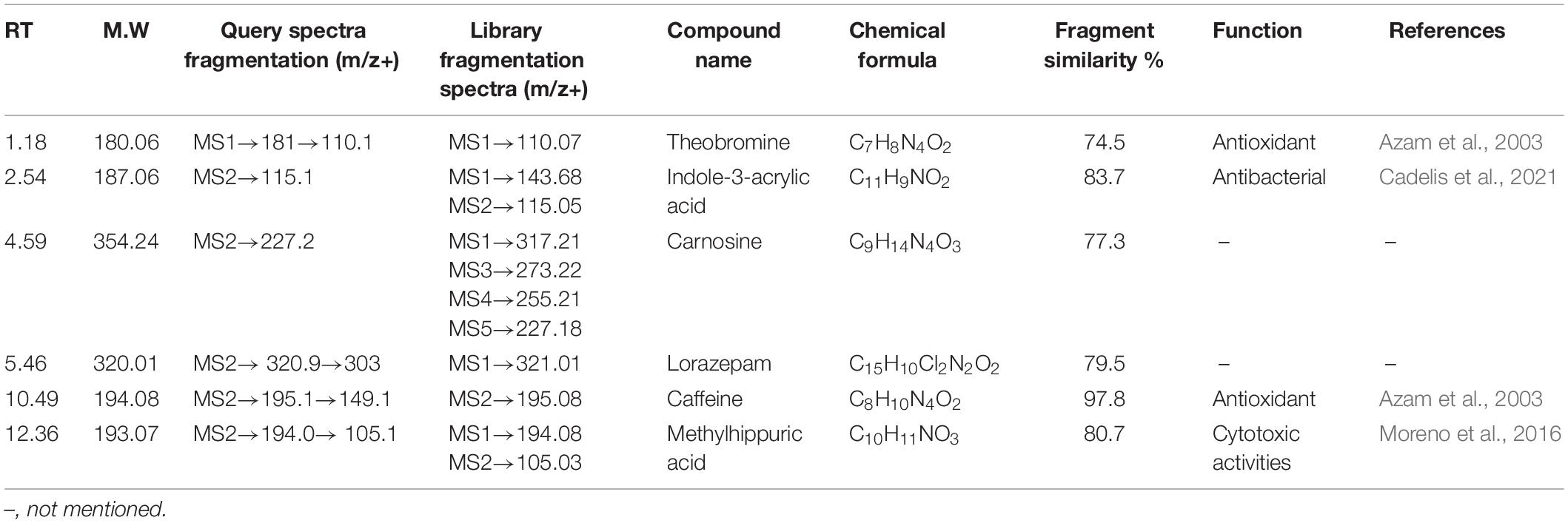
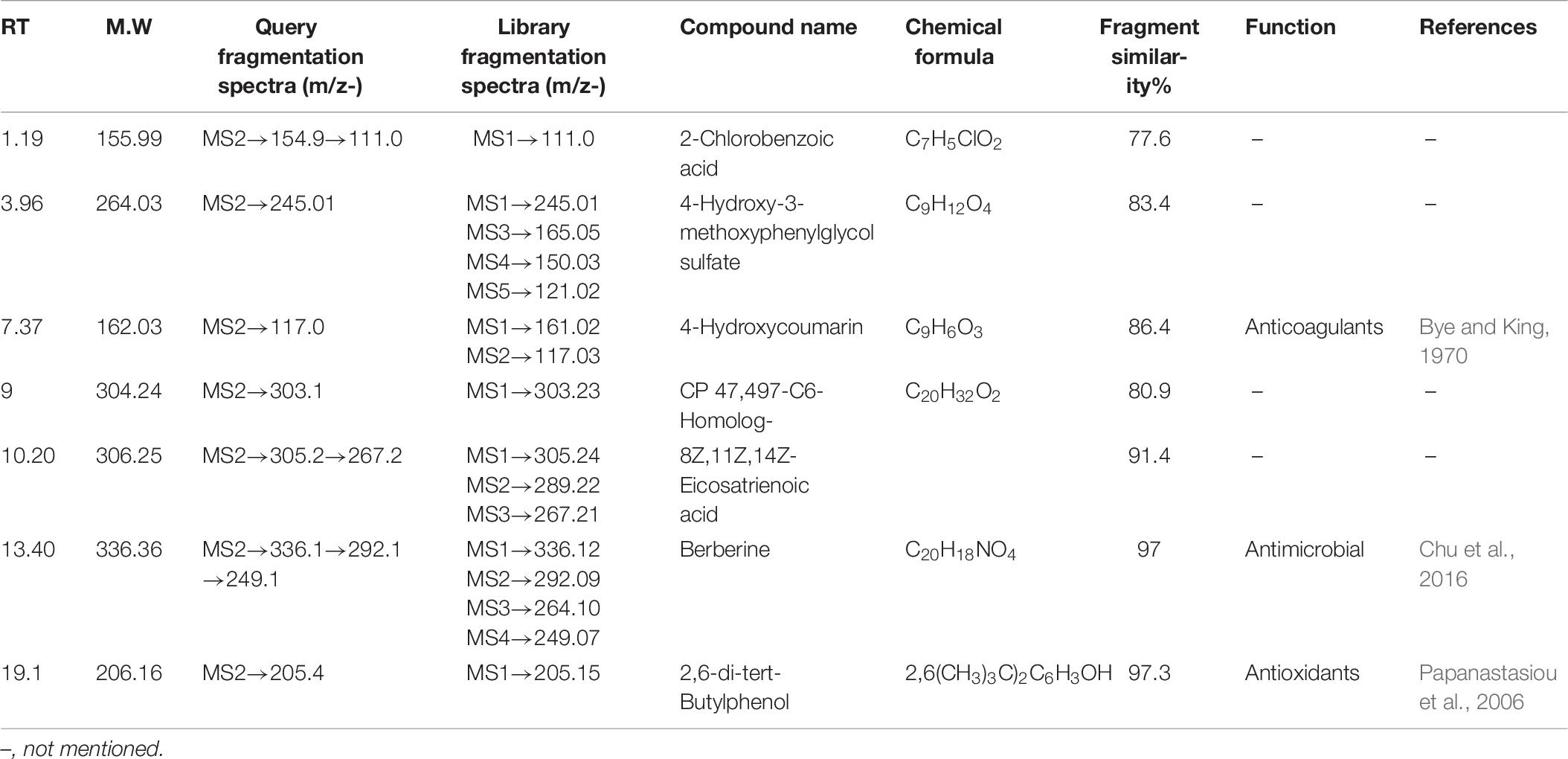
All the compounds were identified by the interpretation of their mass spectral behavior obtained from HPLC-ESI-MS/MS spectra, the accurate mass, and observed MSn fragments that were acquired from ESI+ and ESI– mode and also taking into account data provided by the literature. Raw data of HPLC-ESI-MS/MS were analyzed with ACD/Spectrus software to obtain the top 10 highest m/z values, and these values were subjected to mzCloud Mass Spectral Database for compound identification. The characteristic fragmentation pattern of all components was tentatively elucidated using structurally relevant product ions, and the details of compounds were identified (Tables 5, 6). Overall, 13 additional compounds were induced in the 100 nM BRD4770-treated culture, of which 6 compounds were identified in positive (Figure 4B and Table 5) and 7 in negative (Figure 4D and Table 6) ionization mode of LC-ESI-MS/MS. A total of 14 compounds were detected in both the control and the treated cultures including 1 in positive and 13 in negative mode, respectively (Figure 4 and Supplementary Table 2). However, two compounds, i.e., nimesulide (Supplementary Figure 5A) and N-(2-oxo-3-azepanyl)benzenesulfonamide (Supplementary Figure 5B) of negative mode with RT 6.89 and 11.90, respectively, were not identified in the treated crude extract as compared to the control.
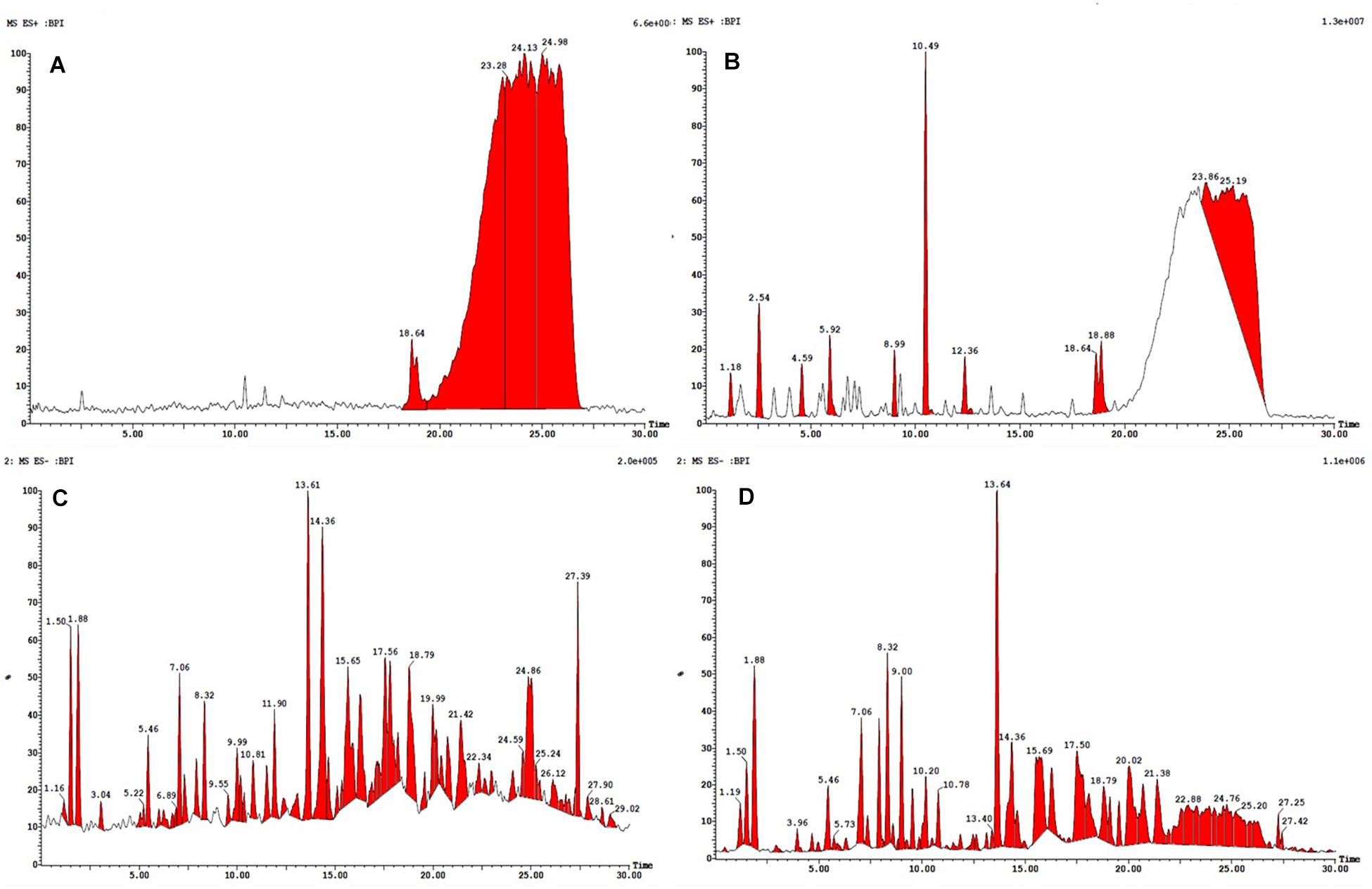
Figure 4. HPLC-ESI-MS/MS base peak chromatogram (BPC) of metabolite (A) non-treated (positive), (B) treated with 100 nM BRD4770 (positive), (C) non-treated (negative), and (D) treated with 100 nM BRD4770 (negative).
HPLC-ESI-MS/MS Identification of Induced Compounds (Positive Ion Mode) in Treated Culture
Theobromine—It showed loss of isocyanic acid (O–C = NH) [M+H–43]+, produced ions at m/z 138.1, further loss of a CO unit (28 Da), and produced ions at m/z 110 (Supplementary Figure 3A; Bianco et al., 2009). Indoleacrylic acid—Major m/z 188.0 after removal of [M+H–72]+ molecular mass probably in the form of CO2 and CO gives rise to m/z 115.1 as a final product ion (Supplementary Figure 3B). Carnosine—Parent m/z 355.2 results from the loss of CO [M+H–28]+ gives 227.2 as a product ion (Supplementary Figure 3C). Lorazepam—Probably by loss of water [M+H–18]+ forms a stable aromatic structure (Supplementary Figure 3D). Caffeine—The protonated molecule of caffeine at m/z 195 gives a major product ion. The ion at m/z 149.1 is the result of a ring contraction with loss of acetaldehyde (CH3CH2OH) [M+H–44]+ (Supplementary Figure 3E; Bianco et al., 2009). Methylhippuric acid—Parent ion m/z 194.0814 after loss of NHCH2COOH [M+H–74]+ and further loss of CH3 [M+H–15]+ gives product m/z 105 (Supplementary Figure 3F; Ohashi et al., 2006). Induced compounds of this positive ion mode are enlisted (Table 5).
HPLC-ESI-MS/MS Identification of Induced Compounds (Negative Ion Mode) in Treated Culture
2-Chlorobenzoic acid—Deprotonated molecule gives m/z 154.9 a major product ion. The ion at m/z 111.0 is the result of loss of CH3CHO [M–H–18] (Supplementary Figure 4A). 4-Hydroxy-3-methoxyphenylglycol sulfate—Parent (m/z) 263.0 resulted to m/z 245 by removal of H2O [M–H–18]– (Supplementary Figure 4B). Hydroxycoumarin—Parent m/z 161 gives m/z 117 product ion after removal of CH3CHO [M–H–44] moieties (Supplementary Figure 4C). CP47, 497-C6-Homolog was found as an m/z 303.1 major product ion (Supplementary Figure 4D). 8Z, 11Z, 14Z-Eicosatrienoic acid—Parent m/z 305.2 after removal of [M–H–38]– gives 267.2 as a product ion (Supplementary Figure 4E). Berberine—Parent (m/z) 336.1 gives m/z 292.1 ion presumably resulting from loss of CH3CHO [M–H–44]– and further m/z 249.1 resulted from loss of COCH3 [M–H–43]– moieties (Wang et al., 2017; Supplementary Figure 4F). 2,6-di-tert-Butylphenol gives m/z 205.4 as a final product of negative ion (Supplementary Figure 4G). Induced compounds of this negative ion mode are also mentioned (Table 6).
RP-HPLC Analysis Result of Major Components, Berberine, Caffeine, and Theobromine
In RP-HPLC, the RT of the standard solution of berberine (RT-10.100) (Figure 5C), caffeine (RT-3.374), and theobromine (RT-6.451) (Figure 5D) was found, showing absorption maxima at 346.5 nm (Figure 6A; Chan et al., 2007), 259.4 nm (Vu et al., 2020; Figure 6C), and 278.4 nm (Caudle et al., 2001; Figure 6E), respectively. According to several literatures, the above compounds show similar absorption spectra at a given wavelength. The RP-HPLC chromatogram of the treated crude compound of D. longicolla showed slight changes in RT as 10.344, 3.478, and 6.058 (Figure 5B) with similar absorption maxima at a given wavelength of 346.5 nm (Figure 6B), 259.4 nm (Figure 6D), and 278.4 nm (Figure 6F), respectively, while RP-HPLC chromatogram of non-treated crude compounds of D. longicolla did not show any peak of the above RT (Figure 5A).
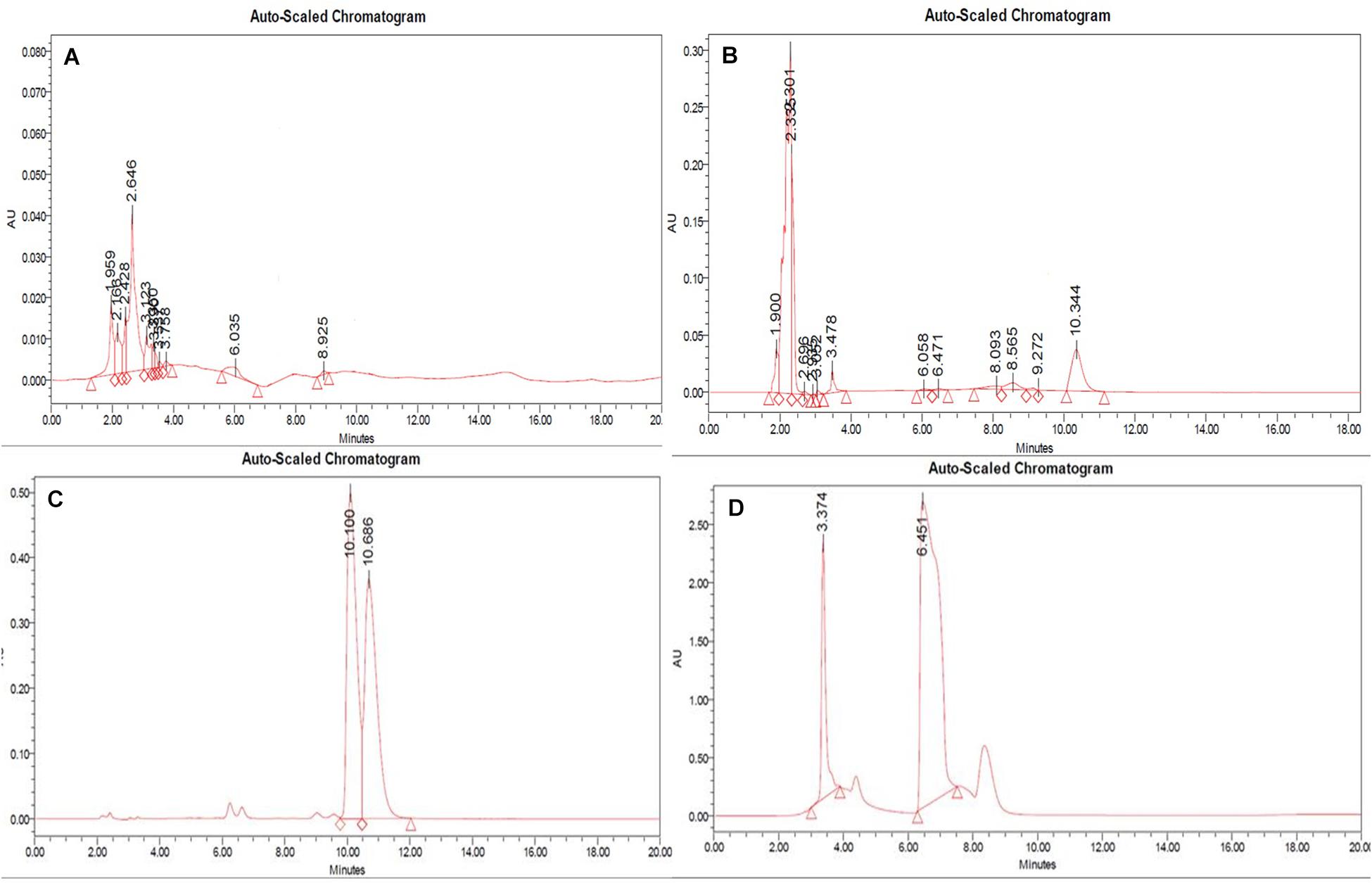
Figure 5. RP-HPLC (A) non-treated, (B) 100 nM BRD4770-treated crude metabolite of D. longicolla, (C) berberine standard, and (D) caffeine + theobromine standard.
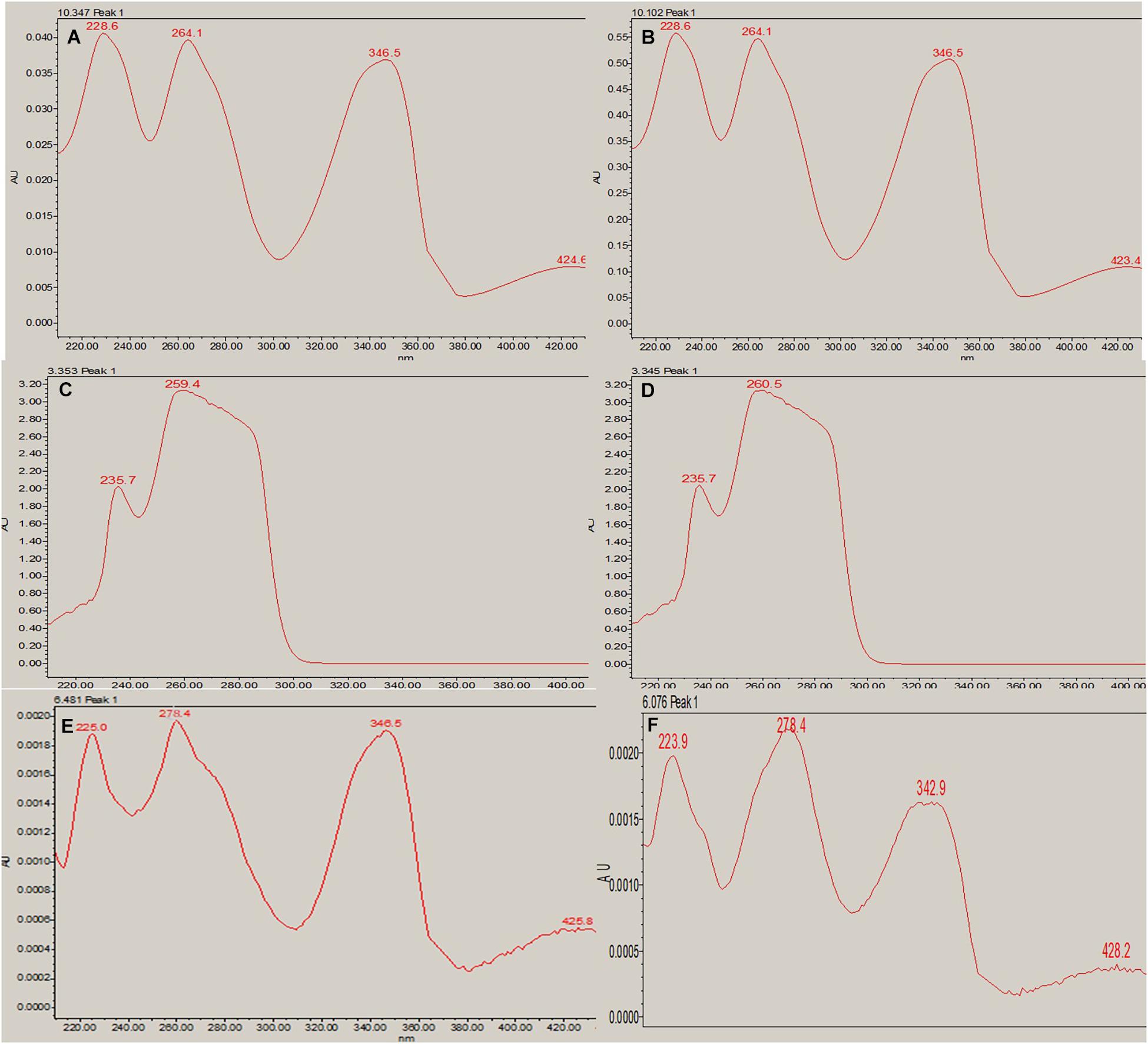
Figure 6. RP-HPLC absorption maxima. (A) Berberine standard at a given wavelength of 346.5 nm (RT-10.347). (B) Berberine in the 100 nM BRD4770-treated crude metabolite at a given wavelength of 346.5 nm (RT-10.102). (C) Caffeine standard at a given wavelength of 259.4 nm (RT-3.353). (D) Caffeine in 100 nM BRD4770-treated crude metabolite at a given wavelength of 260.5 nm (RT-3.345). (E) Theobromine standard at a given wavelength of 278.4 nm (RT-6.451). (F) Theobromine in 100 nM BRD4770-treated crude metabolite at a given wavelength of 278.4 nm (RT-6.076).
Discussion
Endophytic fungus D. longicolla is a commonly reported plant endophyte that has been isolated from a wide range of hosts and is a reliable source for a variety of bioactive compounds (Flores et al., 2013). It has been observed that in fungi, the more novel natural compounds can be obtained, and the production of already known compounds can be up-scaled through an epigenetic regulation if treated with the right epigenetic modifiers in appropriate dose (Zutz et al., 2014). Therefore, the isolate of D. longicolla was grown on PDB medium with different concentrations of BRD4770 to check their epigenetic effect on gene(s) responsible for the production of cryptic metabolites that are not produced under normal culturing conditions.
Epigenetic mechanisms in eukaryotes are highly dynamic phenomena that modulate gene(s) expression regulated through chromatin structure, i.e., mainly controlled by DNA methylation, acetylation, phosphorylation, non-coding RNAs, nucleosome remodeling histone variants, and post-translational modifications (PTMs) of histone. The “histone code” consists of specific patterns of PTMs (post-translational modification) with flexible and charged N-terminus amino acids rich in arginine and lysine residues on histone tails that regulate chromatin compaction and nucleosome positioning in facultative heterochromatin (Lachner et al., 2003). Enzymes (proteins) that have a direct involvement in PTMs of histones include histone-lysine N-methyltransferases (PKMTs) and histone-arginine N-methyltransferases (PRMTs), which catalyze the transfer of one, two, or three methyl groups to lysine and arginine residues of histone proteins. Di- and tri-methylation of lysine 9 on histone H3 (H3K9Me2 and H3K9Me3) mediated by PKMT typically associated with repression (Vastenhouw and Schier, 2012). BRD4770 acts as an inhibitor to H3K9Me2, H3K9Me3, and PKMT. Methylation of H3K9 in humans is controlled by G9a [euchromatic histone-lysine N-methyltransferase 2 (EHMT2)] (Bannister et al., 2001). BRD4770 inhibited G9a with an IC50 of 6.3 μM and was selective for G9a, DNMT (DNA methyl transferase), and HDAC (histone deacetylase) (Kubicek et al., 2007).
In the present study, it was noticed that the crude extract of D. longicolla culture treated with 100 nM BRD4770 performed better in antioxidant and antibacterial assays. The EC50 value of the treated crude extract was much higher compared to that of the non-treated control. Moreover, the antibacterial activity was also increased against MRSA and S. aureus by the respective concentration of the BRD4770-treated crude extract. The findings suggest that the presence of particular concentration of BRD4770 in the growth medium certainly stimulated the secretion of some additional active compounds into the medium, which were not produced under conditions without BRD4770. These compounds could be the reason for increased EC50 for antioxidant activity and enhanced antibacterial activity against tested pathogenic bacteria. It clearly indicates that some of the cryptic (silent) gene(s) or gene clusters get activated and ultimately lead to the production of cryptic metabolites. Methyl ester BRD4770 at 10 μM significantly reduced cellular levels of H3K9me2 and H3K9me3 and increased cellular levels of H3K9me1, and thus, the expression of several new genes were observed in eukaryotes (Yuan et al., 2012). In this study, different concentrations of BRD4770 (1 nM to 1 μM) were used, but only 100 nM was found to be effective for activation of cryptic metabolites in broth medium. GC-MS analysis showed the difference in the crude compound composition present in treated and non-treated cultures. One hundred nanomolar BRD4770-treated GC-MS crude chromatogram clearly indicated that the non-polar compounds (Table 3) were induced. HPLC-ESI-MS/MS analyses showed the difference in crude compound composition in treated and non-treated cultures (Tables 5, 6) that further led to confirmation of the compounds that were induced after treatment with 100 nM BRD4770. Previous reports and literature have shown that among the induced compounds identified by HPLC-ESI-MS/MS, berberine had antibacterial properties against MRSA, additionally known to enhance the inhibitory efficacy of antibiotics (Chu et al., 2016), while caffeine and theobromine have antioxidant activity (Azam et al., 2003). GC-MS and HPLC-ESI-MS/MS were used for the identification of compounds and differences in the crude chromatogram composition present in the treated and non-treated cultures, while RP-HPLC was used to find similar absorption maxima of major compounds like berberine, caffeine, and theobromine in both treated and standard chromatogram at a given wavelength in the RP-HPLC. Similar absorption maxima strongly confirm the presence of these compounds. Induction of berberine may be the reason behind an increased antibacterial activity against MRSA. The antibacterial activity of the crude compound was found only against MRSA and S. aureus while E. coli, S. boydii, and K. pneumoniae were unaffected, probably because of resistance genes present within plasmids of the unaffected bacteria (Radstrom et al., 1991). The treated crude extract had shown increased antioxidant characteristics, which could be justified by the presence of caffeine and theobromine, which certainly were induced due to BRD4770 treatment. Based on the results obtained from this study and the mode of action of BRD4770 mentioned in literature, it is assumed that 100 nM BRD4770, after entering fungal cells, may be involved in the modification of histone as it does inside mammalian cells. Histone modification ultimately leads to upregulation/downregulation of silent genes/or gene clusters in causing the induction and disappearance of compounds present in crude extract. It would be contextual to mention that the additional compounds observed in GC-MS, HPLC-ESI-MS/MS, and RP-HPLC analyses may not represent completely all the additional or novel compounds induced in treated fungus, but it certainly enhances our understanding about the effects of the epigenetic modulator BRD4770 on a microbial system, the endophyte D. longicolla.
Conclusion
The crude metabolite of the endophytic fungus D. longicolla was found to have potent antioxidant and antibacterial activity, which were selected for the treatment of the epigenetic modulator BRD4770. The dose of 100 nM BRD4770 used to treat cultures of the endophytic fungus D. longicolla was noted as an effective concentration in inducing the isolation of bioactive cryptic metabolites, thereby increasing antibacterial and antioxidant activities. A comparative study of BRD4770-treated and non-treated crude chromatograms of RP-HPLC with standard solutions of berberine, caffeine, and theobromine confirms the presence of respective compounds in treated cultures. This study successfully establishes the importance of BRD4770, which also interacts with epigenetic targets and can significantly induce and downregulate the production of cryptic metabolites in the endophytic fungus D. longicolla. The histone level modification brought out by the treatment of BRD4770 will further help to explore the exact mechanisms/pathways involved.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author Contributions
JN, RK, and AS hypothesized the research work, wrote the manuscript, analyzed the data, and acquired funding. JN, PP, and RB performed the research. VS, AS, and VG analyzed the data. RK reviewed the manuscript and supervised the research. All authors contributed to the article and approved the submitted version.
Funding
JN wishes to thank the Department of Biotechnology (DBT) (award no.: DBT/JRF/BET-16/I/2016/AL/135), New Delhi, India, for financial support. RK appreciates the SERB, DST, New Delhi (EEQ/2020/000549) and IoE (R/Dev/D/IoE/Incentive/2021-22/32181), BHU, Varanasi, for financial help.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors are thankful to the Head and Coordinator, CAS and DST-FIST in Botany, Institute of Science, BHU, Varanasi, India, for providing essential research facilities. JN, AS, PP, and RB are thankful to DBT, UGC, and CSIR, respectively, for providing the JRFs and SRFs. The authors appreciably acknowledge the help of ISLS, UGC-UPE, DST-PURSE, BHU, Varanasi, India, for HPLC and GC-MS analysis and Gopal Nath, Institute of Medical Sciences, BHU, Varanasi, India, for antibacterial assay facility.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.725463/full#supplementary-material
References
Agusta, A., Ohashi, K., and Shibuya, H. (2006). Bisanthraquinone metabolites produced by the endophytic fungus Diaporthe sp. Chem. Pharm. Bull. 54, 579–582. doi: 10.1248/cpb.54.579
Al-Dhabi, N. A., Esmail, G. A., Duraipandiyan, V., and Arasu, M. V. (2019). Chemical profiling of Streptomyces sp. Al-Dhabi-2 recovered from an extreme environment in Saudi Arabia as a novel drug source for medical and industrial applications. Saudi J. Biol. Sci. 26, 758–766. doi: 10.1016/j.sjbs.2019.03.009
Arnold, A. E., and Lutzoni, F. (2007). Diversity and host range of foliar fungal endophytes: are tropical leaves biodiversity hotspots. Ecology 88, 541–549. doi: 10.1890/05-1459
Azam, S., Hadi, N., Khan, N. U., and Hadi, S. M. (2003). Antioxidant and prooxidant properties of caffeine, theobromine and xanthine. Med. Sci. Monit. 9, BR325–BR330.
Bannister, A. J., Zegerman, P., Partridge, J. F., Miska, E. A., Thomas, J. O., Allshire, R. C., et al. (2001). Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410, 120–124. doi: 10.1038/35065138
Bianco, G., Novario, G., Zianni, R., and Cataldi, T. R. (2009). Comparison of two SPME fibers for the extraction of some off-flavor cork-taint compounds in bottled wines investigated by GC–HRMS. Anal. Bioanal. Chem. 393, 2019–2027. doi: 10.1007/s00216-009-2632-0
Bubák, F. and Kabát, J. E. (1905). Vierter Beitrag zur Pilzflora von Tirol. Öst. Bot. Z. 55, 73–79. doi: 10.1007/bf01791128
Bye, A., and King, H. K. (1970). The biosynthesis of 4-hydroxycoumarin and dicoumarol by Aspergillus fumigatus Fresenius. Biochem. J. 117, 237–245. doi: 10.1042/bj1170237
Cadelis, M. M., Li, S. A., Bourguet−Kondracki, M. L., Blanchet, M., Douafer, H., Brunel, J. M., et al. (2021). Spermine Derivatives of Indole−3−carboxylic Acid, Indole−3−acetic Acid and Indole−3−acrylic Acid as Gram−Negative Antibiotic Adjuvants. Chem. Med. Chem. 16, 513–552. doi: 10.1002/cmdc.202000359
Caudle, A. G., Gu, Y., and Bell, L. N. (2001). Improved analysis of theobromine and caffeine in chocolate food products formulated with cocoa powder. Food Res. Int. 34, 599–603. doi: 10.1016/s0963-9969(01)00077-1
Chan, C. O., Chu, C. C., Mok, D. K. W., and Chau, F. T. (2007). Analysis of berberine and total alkaloid content in Cortex Phellodendri by near infrared spectroscopy (NIRS) compared with high-performance liquid chromatography coupled with ultra-visible spectrometric detection. Anal. Chim. Acta 592, 121–131. doi: 10.1016/j.aca.2007.04.016
Chu, M., Zhang, M. B., Liu, Y. C., Kang, J. R., Chu, Z. Y., Yin, K. L., et al. (2016). Role of berberine in the treatment of methicillin-resistant Staphylococcus aureus infections. Sci. Rep. 6:24748.
de Bary, A. (1866). Morphologie Und Physiologie Der Pilze, Flechten und Myxomyceten. Germany: Wilhelm Engelmann.
Flores, A. C., Pamphile, J. A., Sarragiotto, M. H., and Clemente, E. (2013). Production of 3-nitropropionic acid by endophytic fungus Phomopsis longicolla isolated from Trichilia elegans A. JUSS ssp. elegans and evaluation of biological activity. World J. Microbiol. Biotechnol. 29, 923–932. doi: 10.1007/s11274-013-1251-2
Foo, L. W., Salleh, E., and Hana, S. N. (2017). Green extraction of antimicrobial bioactive compound from piper betle leaves: probe type ultrasound-assisted extraction vs supercritical carbon dioxide extraction. Chem. Eng. Trans. 56, 109–114.
Kharwar, R. N., Mishra, A., Gond, S. K., Stierle, A., and Stierle, D. (2011). Anticancer compounds derived from fungal endophytes: their importance and future challenges. Nat. Prod. Rep. 28, 1208–1228. doi: 10.1039/c1np00008j
Kharwar, R. N., Sharma, V. K., Mishra, A., Kumar, J., Singh, D. K., Verma, S. K., et al. (2020). Harnessing the Phytotherapeutic Treasure Troves of the Ancient Medicinal Plant Azadirachta indica (Neem) and Associated Endophytic Microorganisms. Planta. Med. 86, 906–940. doi: 10.1055/a-1107-9370
Kharwar, R. N., Verma, V. C., Strobel, G., and Ezra, D. (2008). The endophytic fungal complex of Catharanthus roseus (L.) G. Don. Curr. Sci. 95, 228–233.
Kubicek, S., O’Sullivan, R. J., August, E. M., Hickey, E. R., Zhang, Q., Teodoro, M. L., et al. (2007). Reversal of H3K9me2 by a small-molecule inhibitor for the G9a histone methyltransferase. Mol. Cell 25, 473–481. doi: 10.1016/j.molcel.2007.01.017
Lachner, M., O’Sullivan, R. J., and Jenuwein, T. (2003). An epigenetic road map for histone lysine methylation. J. Cell Sci. 116, 2117–2124. doi: 10.1242/jcs.00493
Meeran, S. M., Ahmed, A., and Tollefsbol, T. O. (2010). Epigenetic targets of bioactive dietary components for cancer prevention and therapy. Clin Epigenet. 1, 101–116. doi: 10.1007/s13148-010-0011-5
Miller, N. J., and Rice-Evans, C. A. (1997). Factors influencing the antioxidant activity determined by the ABTS+ radical cation assay. Free Radic. Res. 26, 195–199. doi: 10.3109/10715769709097799
Moreno, P. G. G., Wang, L. Y., Alvarez, M. R., Olivar, D. D. B. D., Echavez, M. D. J., Yu, G. F. B., et al. (2016). Preliminary Comparative Chemical Profiles and Cytotoxic Activities of Two Philippine Allium sativum Linn. Varieties: ilocos White and Native. PJHRD 20, 1–10. doi: 10.1007/978-0-387-70638-2_84
Naeim, H., El-Hawiet, A., Rahman, R. A. A., Hussein, A., El Demellawy, M. A., and Embaby, A. M. (2020). Antibacterial activity of Centaurea pumilio L. root and aerial part extracts against some multidrug resistant bacteria. BMC Complement Altern. Med. Ther. 20:79. doi: 10.1186/s12906-020-2876-y
Ohashi, Y., Mamiya, T., Mitani, K., Wang, B., Takigawa, T., Kira, S., et al. (2006). Simultaneous determination of urinary hippuric acid, o-, m-and p-methylhippuric acids, mandelic acid and phenylglyoxylic acid for biomonitoring of volatile organic compounds by gas chromatography–mass spectrometry. Anal. Chim. Acta 566, 167–171. doi: 10.1016/j.aca.2006.03.018
Ola, A. R., Debbab, A., Kurtán, T., Brötz-Oesterhelt, H., Aly, A. H., and Proksch, P. (2014). Dihydroanthracenone metabolites from the endophytic fungus Diaporthe melonis isolated from Annona squamosa. Tetrahedron Lett. 55, 3147–3150. doi: 10.1016/j.tetlet.2014.03.110
Papanastasiou, M., McMahon, A. W., Allen, N. S., Doyle, A. M., Johnson, B. J., and Keck-Antoine, K. (2006). The hydrolysis mechanism of bis (2, 4-di-tert-butyl) pentaerythritol diphosphite (Alkanox P24): an atmospheric pressure photoionisation mass spectrometric study. Polym. Degrad. Stab. 91, 2675–2682. doi: 10.1016/j.polymdegradstab.2006.04.023
Petrini, O., Fisher, P. J., and Petrini, L. E. (1992). Fungal endophytes of bracken (Pteridium aquilinum) with some reflections on their use in biological control. Petrini1, PJ Fisher2 and LE PetrinP1. Sydowia 44, 282–293.
Radstrom, P., Swedberg, G. O. T. E., and Sköld, O. (1991). Genetic analyses of sulfonamide resistance and its dissemination in gram-negative bacteria illustrate new aspects of R plasmid evolution. Antimicrob. Agents Chemother. 35, 1840–1848. doi: 10.1128/aac.35.9.1840
Rutledge, P. J., and Challis, G. L. (2015). Discovery of microbial natural products by activation of silent biosynthetic gene clusters. Nat. Rev. Microbiol. 13, 509–523. doi: 10.1038/nrmicro3496
Sharma, V. K., Kumar, J., Singh, D. K., Mishra, A., Verma, S. K., Gond, S. K., et al. (2017). Induction of Cryptic and Bioactive Metabolites through Natural Dietary Components in an Endophytic Fungus Colletotrichum gloeosporioides (Penz.) Sacc. Front. Microbiol. 8:1126. doi: 10.3389/fmicb.2017.01126
Singh, A., Singh, D. K., Kharwar, R. N., White, J. F., and Gond, S. K. (2021). Fungal Endophytes as Efficient Sources of Plant-Derived Bioactive Compounds and Their Prospective Applications in Natural Product Drug Discovery: insights, Avenues, and Challenges. Microorganisms 9:197. doi: 10.3390/microorganisms9010197
Sisin, N. N. T., Suláin, M. D., and Abdullah, H. (2017). Antiproliferative, antioxidative and compounds identification from methanolic extract of passiflora foetida and its fractions. J. Anal. Pharm. Res. 6, 1–10. doi: 10.9734/bjmmr/2016/23767
Vastenhouw, N. L., and Schier, A. F. (2012). Bivalent histone modifications in early embryogenesis. Curr. Opin. Cell Biol. 24, 374–386. doi: 10.1016/j.ceb.2012.03.009
Verma, S. K., Gond, S. K., Mishra, A., Sharma, V. K., Kumar, J., Singh, D. K., et al. (2014). Impact of environmental variables on the isolation, diversity and antibacterial activity of endophytic fungal communities from Madhuca indica Gmel. at different locations in India. Ann. Microbiol. 64, 721–734. doi: 10.1007/s13213-013-0707-9
Verma, V. C., Kharwar, R. N., and Strobel, G. A. (2009). Chemical and functional diversity of natural products from plant associated endophytic fungi. Nat. Prod. Comm. 4, 1511–1532.
Viszwapriya, D., Prithika, U., Deebika, S., Balamurugan, K., and Pandian, S. K. (2016). In vitro and in vivo antibiofilm potential of 2, 4-Di-tert-butylphenol from seaweed surface associated bacterium Bacillus subtilis against group A streptococcus. Microbiol. Res. 191, 19–31. doi: 10.1016/j.micres.2016.05.010
Vu, D. H., Truong Thi, Thu, H., Dong Thi, Ha, L., and Nguyen Mai, H. (2020). RP-HPLC and UV Spectrophotometric Analysis of Paracetamol, Ibuprofen, and Caffeine in Solid Pharmaceutical Dosage Forms by Derivative, Fourier, and Wavelet Transforms: a Comparison Study. J Anal Methods Chem 2020:8107571.
Wang, K., Feng, X., Chai, L., Cao, S., and Qiu, F. (2017). The metabolism of berberine and its contribution to the pharmacological effects. Drug Metab. Rev. 49, 139–157. doi: 10.1080/03602532.2017.1306544
Yuan, Z., Li, J., Li, J., Zhang, L., Gao, X., Gao, H. J., et al. (2012). Investigation on BRCA1 SNPs and its effects on mastitis in Chinese commercial cattle. Gene 505, 190–194. doi: 10.1016/j.gene.2012.05.010
Keywords: endophytic fungus, epigenetic modulator, GC-MS, LC-ESI-MS/MS, reverse-phase HPLC
Citation: Nishad JH, Singh A, Bharti R, Prajapati P, Sharma VK, Gupta VK and Kharwar RN (2021) Effect of the Histone Methyltransferase Specific Probe BRD4770 on Metabolic Profiling of the Endophytic Fungus Diaporthe longicolla. Front. Microbiol. 12:725463. doi: 10.3389/fmicb.2021.725463
Received: 15 June 2021; Accepted: 17 August 2021;
Published: 17 September 2021.
Edited by:
Patrick Eichenberger, New York University, United StatesReviewed by:
Ekachai Chukeatirote, Mae Fah Luang University, ThailandDebdulal Banerjee, Vidyasagar University, India
Copyright © 2021 Nishad, Singh, Bharti, Prajapati, Sharma, Gupta and Kharwar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ravindra Nath Kharwar, rnkharwar@gmail.com; rnkharwarbot@bhu.ac.in
 Jay Hind Nishad
Jay Hind Nishad Arti Singh
Arti Singh Rajnish Bharti
Rajnish Bharti Priyanka Prajapati
Priyanka Prajapati Vijay Kumar Sharma
Vijay Kumar Sharma Vijai Kumar Gupta
Vijai Kumar Gupta Ravindra Nath Kharwar
Ravindra Nath Kharwar