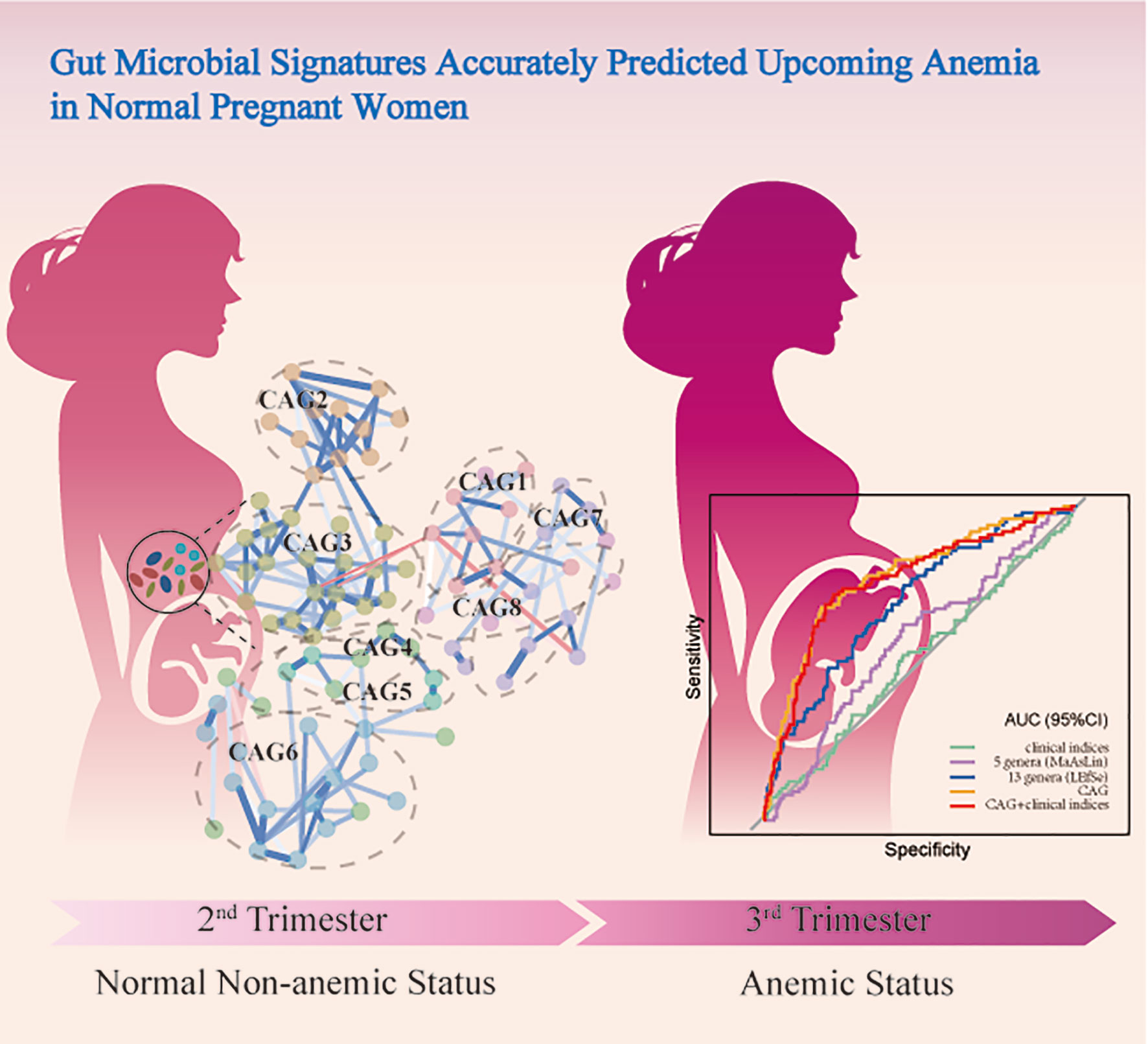
Insight Into the Potential Value of Gut Microbial Signatures for Prediction of Gestational Anemia
- 1State Key Laboratory of Reproductive Medicine, School of Public Health, Nanjing Medical University, Nanjing, China
- 2Key Laboratory of Modern Toxicology of Ministry of Education, School of Public Health, Nanjing Medical University, Nanjing, China
- 3Nanjing Maternity and Child Health Care Institute, Women’s Hospital of Nanjing Medical University, Nanjing Maternity and Child Health Care Hospital, Nanjing, China
- 4Department of Endocrinology, Children’s Hospital of Nanjing Medical University, Nanjing, China
The gut microbiota alternations are associated with gestational anemia (GA); however, limited predictive value for the subsequent incidence of anemia in normal gestational women has been obtained. We sought to rigorously characterise gut dysbiosis in subjects with GA and explored the potential predictive value of novel microbial signatures for the risk of developing GA. A prospective cohort of subjects with GA (n = 156) and healthy control (n = 402), all of whom were free of GA in the second trimester, by 16S rRNA gene sequencing was conducted. Microbial signatures altered dramatically in GA compared with healthy control in the second trimester. Megamonas, Veillonella, and Haemophilus were confirmed to show differential abundances in GA after adjusting for covariates. On the contrary, Lachnospiraceae and Blautia were enriched in control. Microbial co-abundance group (CAG) network was constructed. Prospectively, CAG network relatively accurately predicted upcoming GA in normal pregnant women with an AUC of 0.7738 (95%CI: 0.7171, 0.8306) and the performance was further validated in Validation set (0.8223, 95%CI: 0.7573, 0.8874). Overall, our study demonstrated that alterations in the gut microbial community were associated with anemia in pregnancy and microbial signatures could accurately predict the subsequent incidence of anemia in normal pregnant women. Our findings provided new insights into understanding the role of gut microbiota in GA, identifying high-risk individuals, and modulating gut microbiota as a therapeutic target, thus improving quality of life and well-being of women and children.
Introduction
Anemia is a serious public health problem affecting people all over the world, and is one of the most frequent complications involved in pregnancy, imposing a tremendous toll on well-being of approximately 40% of pregnant women in China (Li et al., 2018). Gestational anemia, according to the World Health Organization (WHO), is defined as a hemoglobin concentration (Hb) < 110 g L-1. Gestational anemia mostly occurred in the second and third trimester (Zhao et al., 2018). Multiple factors account for gestational anemia, nutritional iron deficiency anemia (IDA) (approximately 75%) and folate megaloblastic deficiency anemia being the commonest (Sifakis and Pharmakides, 2000; Goonewardene et al., 2012).
Anemia in pregnancy has adverse effects on maternal and neonatal health. Weakness, fatigue, being vulnerable to infection, reduced work capacity, and productivity are typical symptoms during pregnancy (Prema et al., 1982; Hunt, 2002). Current evidence suggests severe gestational anemia could be associated with an increased risk of preterm birth, low birth weight, and even neonatal and maternal mortality (Khan et al., 2006; Rahman et al., 2016; Figueiredo et al., 2018; Guignard et al., 2021). Furthermore, it has been reported that babies born to anemic mothers are prone to exhibit future poor cognitive performance and delayed mental and motor development in adolescence and adulthood (Anker et al., 2009; Camaschella, 2015).
The current clinical method is generally applied to cross-sectional diagnosis rather than prediction in the long term. Although Hb concentration serves as a golden standard for the diagnosis of anemia in pregnancy, it has limitations in precise prediction of potential impending anemia in normal pregnant women. Additionally, it remains controversial to use Hb concentration to distinguish true or absolute anemia from relative anemia, ascribed to a normal physiologic increase of plasma volume (Sifakis and Pharmakides, 2000). Thus, an accurate determination of gestational anemia is essential and of enormous clinical importance for prevention and management of anemia in pregnancy.
The human gut hosts an immense number of resident microorganisms, collectively termed as the microbiota (Structure, Function and Diversity of the Healthy Human Microbiome, 2012; Bäckhed et al., 2004). There has been accumulating evidence that the gut microbiota is implicated in enteric, metabolic, and psychiatric diseases (Imhann et al., 2018; Vich Vila et al., 2018; Thomas et al., 2019; Wirbel et al., 2019). Previous studies have documented that gut microbial dysfunction (e.g., deficiency in lactobacillus) is related to IDA and gut microbiota could promote hematopoiesis, underlying the close relationship between gut microbiota and anemia (Balamurugan et al., 2010; Khosravi et al., 2014). During pregnancy, host profoundly remodeling of the gut microbiota could cause symptoms of metabolic syndrome, which might be related to transportation and storage of host Fe, and ultimately lead to the occurrence of gestational anemia (Koren et al., 2012). Microbial signatures have been elucidated to function as novel biomarkers to discriminate patients suffering from illness and healthy individuals (Liu et al., 2019; Wei et al., 2020). To date, an anemia classifier in pregnancy has been constructed (Long et al., 2021); however, no efforts have been made to predict the subsequent incidence of anemia in normal gestational women.
To reach a better understanding of how gestational anemia is developing and regulated, herein, we conducted a prospective study to rigorously evaluate dynamic landscape of gut microbiota varying from healthy status to identified anemic status in pregnant women. Leveraging the discriminative microbial signatures in the early stage of pregnancy when anemia did not occur yet, we prospectively built an accurate prediction model for the subsequent incidence of gestational anemia. Our findings help identify pregnant women with high risk of anemia in general population and ultimately improve quality of life and well-being of women and children.
Materials and Methods
Study Design and Participants
This study used data from the Mother and Child Microbiome Cohort (MCMC) Study, a prospective birth cohort study initiated and maintained in Nanjing Maternity and Child Health Care Hospital. The recruitment of eligible study pregnant women was from 2017 to 2018 (n = 1527), when they were all in the second trimester. This cohort aimed to explore the relationship between gut microbiota and maternal and children’s health.
Among 1527 eligible women who were enrolled, 781 subjects have provided fecal samples both in the second and third trimester. Then, those with assisted conception (n = 19) or twin pregnancy (n = 8) were excluded. Additionally, women with pregnant complications containing diabetes mellitus and pathological anemia (e.g., aplastic anemia and hemolytic anemia) were excluded (n = 142). Women with diagnosed anemia in the second trimester were excluded (n = 54). In the final analysis, 558 participants were included (Supplementary Figure 1).
Though Hb < 110 g L-1 has been the accepted criterion for the diagnosis of gestational anemia, we reduced the diagnostic level to Hb < 100 g L-1 considering the increasing plasma volume during pregnancy, to reduce bias (Lund and Donovan, 1967). All subjects accepted treatment after they were diagnosed with anemia in our cohort.
Ethics
We obtained signed informed consent from all participants. The study was approved by the Ethics Committee of Nanjing Medical University [FWA00001501 No. (2017) 003].
Sample Collection and Sequencing Data
All samples were collected prior to anemia treatment. Fecal samples were obtained from each participant for measuring gut microbiota during the second (at 24 weeks of pregnancy) and third trimester (at 32 weeks of pregnancy), respectively, and were stored at -80°C until DNA extraction. Serum samples were obtained during the second trimester and were frozen in -20°C freezers. Serum ferritin levels were measured by electrochemiluminescence immunoassay on the immunoassay analyzer (Beckman Coulter Inc., Fullerton CA, USA) with the same batch of reagents.
16S rRNA sequencing was performed using primers (338F: 5’-ACTCCTACGGGAGGCAGCAG-3’ and 806R: 5’-GGACTACHVGGGTWTCTAAT-3’). Each sample genomic DNA was extracted using QIAamp Fast DNA Stool Mini Kit (Qiagen, Germany). From extracted DNA, we sequenced the hypervariable region 16S rRNA gene V3 to V4 regions by HiSeq2500 PE250 platform in Meiji Bioinformatics Technology Co. Ltd (Nanjing, China). Blinded positive quality control (QC) specimens were used across all sequencing batches for quality control.
The 16S rRNA sequencing data were analyzed using Quantitative Insights Into Microbial Ecology (QIIME2 V.2020.6). The dada2 plugin was used to denoise sequences, and this quality control process will additionally filter any phiX reads (commonly present in marker gene Illumina sequence data) that were identified in the sequencing data, and will filter chimeric sequences. Amplicon sequence variants (ASVs) were obtained at 100% sequence homology; the taxonomy was assigned against the Silva database (Silva 138 release). To minimize the effect of spurious sequences, one case with too low sequence number was excluded. Representative sequences for each ASV were built into a phylogenetic tree with FastTree plugin. Alpha and beta diversity analyzes were conducted at a rarefied sampling depth of 31291.
Statistical Analysis
To compare maternal anemia information by characteristics, t-test for continuous variables and χ² test for categorical variables were used. Multivariate linear regression model was applied to multiple-factor analysis. Model 1 was crude model; Model 2 was adjusted for maternal age, pre pregnant body mass index (BMI), parity, gravidity, family income, maternal education level, passive smoking, antibiotic use during pregnancy, folic acid and iron supplement use during pregnancy, alcohol and caffeine use pre pregnancy, and alcohol and caffeine use during pregnancy.
Wilcoxon rank-sum test was used to compare α-diversity, and permutational multivariate analysis of variance (PERMANOVA) using 9999 permutations was used to test for statistical significance between two groups. Given a false discovery rate (FDR) of 5%, linear discriminant analysis effect size (LEfSe) was used to identify bacteria differentially abundant between anemic women and normal women (Segata et al., 2011). To further validate the results, after adjusting for maternal age, pre pregnant BMI, parity, gravidity, family income, maternal education level, passive smoking, antibiotic use during pregnancy, folic acid and iron supplement use during pregnancy, alcohol and caffeine use pre pregnancy, and alcohol and caffeine use during pregnancy, multivariate association with linear models algorithm (MaAsLin) analysis was performed (Morgan et al., 2012; Wei et al., 2020).
The top 99 most abundant genera were used to construct co-abundance group (CAG) network. Kendall correlation was calculated by the function “cor”. CAG was defined with a Spearman correlation distance metric using the “Made4” R package. The appropriate number of clustering was selected based on significance testing among each group of the original Kendall correlation matrix using “adonis” function in “Vegan” R package. The sum of relative abundance of ASVs which belonged to the same CAG was calculated to represent the abundance of this CAG (Zhang et al., 2021). Subsequently, we used “qgraph” R package to construct the regularized partial correlation network based on least absolute shrinkage and selection operator (lasso) (Epskamp and Fried, 2018; Liang et al., 2020).
Spearman correlation was used to investigate relationships among continuous variables, and point biserial correlation was used to examine relationships between binary variable and continuous variables. Finally, we constructed receiving operational curve (ROC) and calculated area under curve (AUC) to assess the predictive performance of the model with the “pROC” R package.
Results
Characteristics of the Study Population
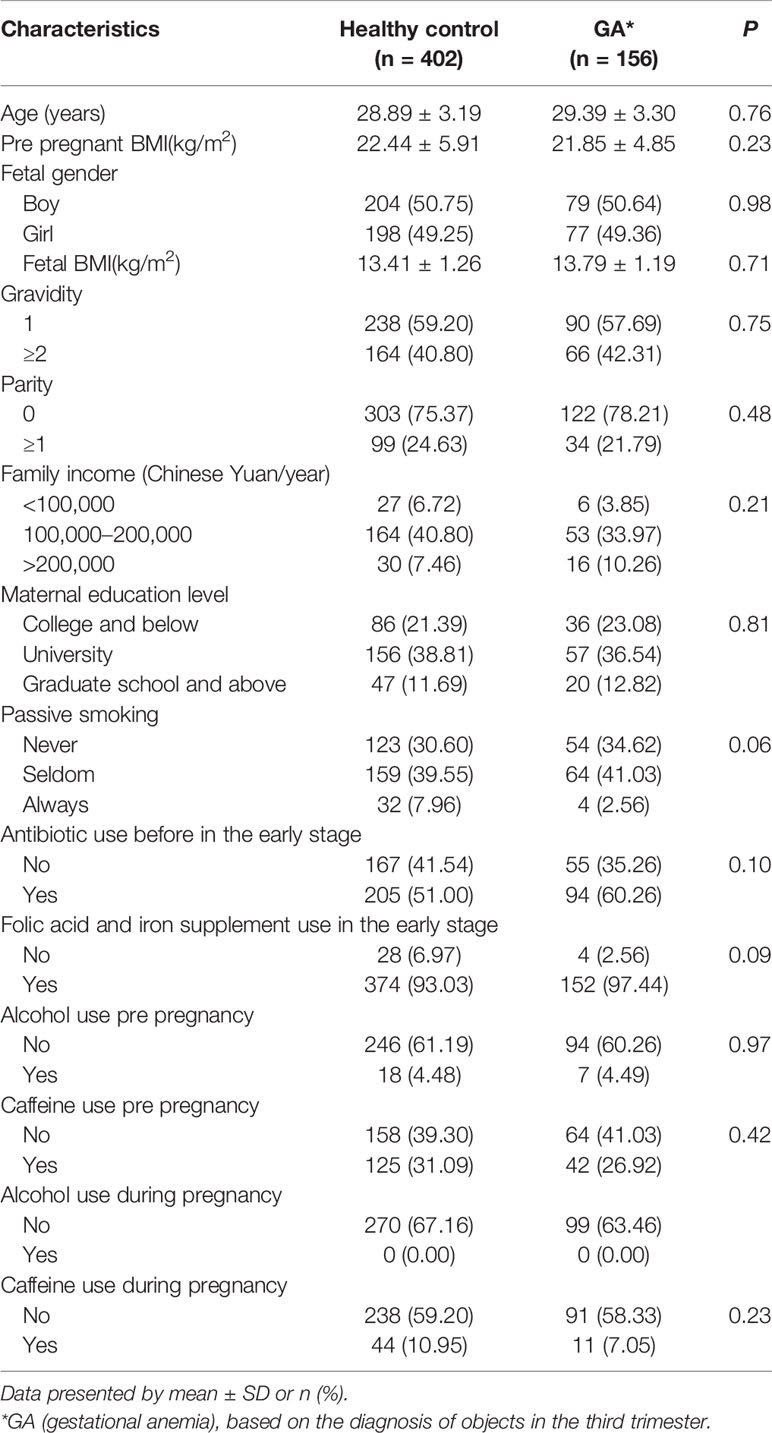
Totally, 156 (27.96%) women diagnosed with gestational anemia (GA) and 402 healthy control in the third trimester, all of whom were non-anemic in the second trimester, constituted the study population for final analysis. Detailed demographic features of the cohort were summarized in Table 1.
Fecal Microbiota Altered Dramatically During Pregnancy
Shannon index indicated progression of pregnancy from the second trimester (T2) to the third trimester (T3) was accompanied by an increment in α-diversity (GA: P < 0.001, Control: P < 0.001, Figures 1A, B) and the observed ASVs suggested the same trend (Supplementary Table 1). Principal coordinate analysis (PCoA) based on unweighted UniFrac distance was conducted to elaborate the overall structure of microbial composition. PERMANOVA manifested significant differences in structure and composition of the microbiota between T2 and T3 (GA: P = 0.001, Control: P = 0.001, Figures 1C, D). Such significant difference was also observed based on weighted UniFrac distance (Supplementary Table 1). LEfSe analysis revealed that remarkable differences were observed between T2 and T3 in pooled group (LDA > 2, FDR < 0.05, Figure 1E and Supplementary Table 2).
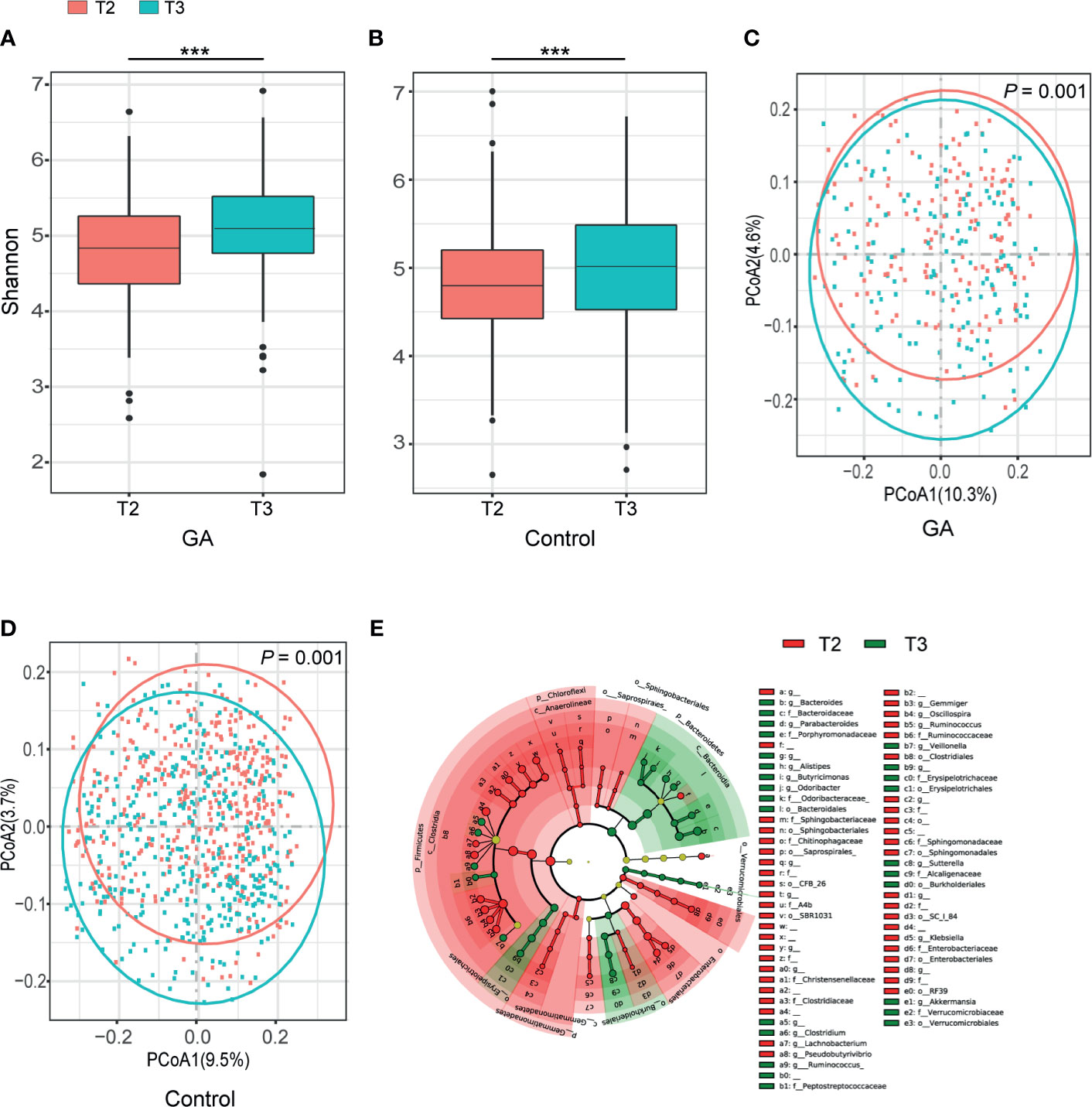
Figure 1 Comparisons of alpha-diversity, beta-diversity, and variations of gut microbiota composition between T2 and T3. (A) Box plots based on Shannon diversity index in GA group. (B) Box plots based on Shannon diversity index in healthy control group. (C) PCoA based on unweighted UniFrac matrix of GA group. (D) PCoA based on unweighted UniFrac matrix of healthy control group. (E) The cladogram showed differently enriched taxa in the second trimester and the third trimester (FDR < 0.05). FDR, false discovery rate; PCoA, principal coordinate analysis; GA, gestational anemia; T2, the second trimester; T3, the third trimester; ***P < 0.001.
Changes in the Gut Microbiota Between GA and Healthy Control
Both Shannon index (T2: P = 0.75, T3: P = 0.06, Figures 2A, B) and the observed ASVs (Supplementary Table 1) in different trimesters showed that there was no difference in richness and diversity of the gut microbiota between GA and healthy control. No significant difference was observed between GA and healthy control based on unweighted or weighted UniFrac distance in both T2 and T3 (T2: P = 0.80, T3: P = 0.15, Figures 2C, D and Supplementary Table 1).
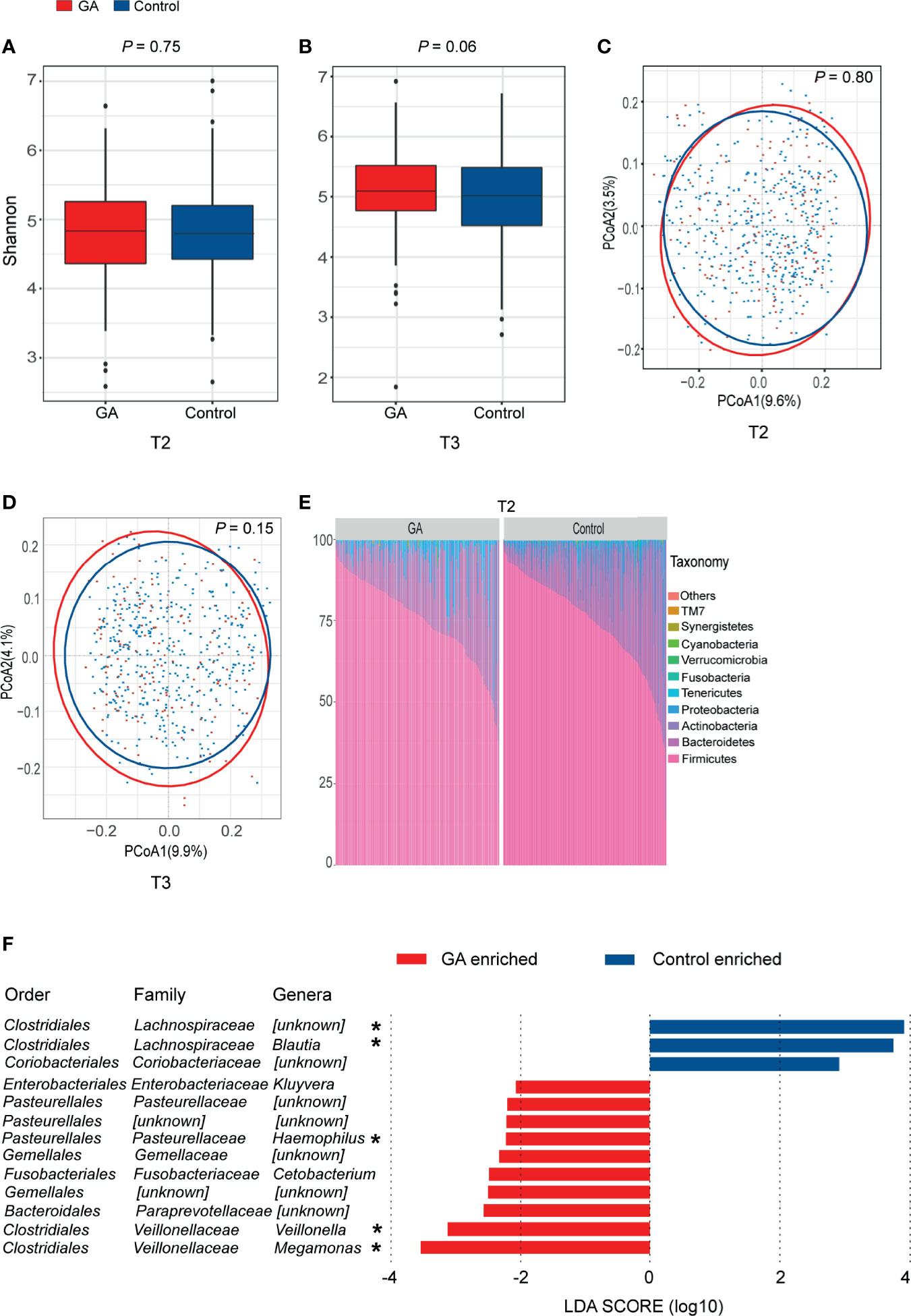
Figure 2 Comparisons of alpha-diversity, beta-diversity, and variations of gut microbiota composition between GA and healthy control. (A) Box plots based on Shannon diversity index in the second trimester. (B) Box plots based on Shannon diversity index in the third trimester. (C) PCoA based on unweighted UniFrac matrix of T2 group. (D) PCoA based on unweighted UniFrac matrix of T3 group. (E) Relative proportions of bacterial phyla in GA and healthy control in the second trimester. (F) Histogram of the LDA scores computed for differentially abundant taxa between GA and healthy control in the second trimester (FDR < 0.05). FDR, false discovery rate; PCoA, principal coordinate analysis; GA, gestational anemia; T2, the second trimester; T3, the third trimester; LDA, linear discriminant analysis; *Genera remained significantly associated with GA after adjusting for covariates using multivariate association with linear models algorithm (MaAsLin). *P < 0.05.
In both T2 and T3, overall microbiota composition of maternal fecal microbiota at phylum level showed that the maternal fecal microbiota was dominated by Firmicutes and Bacteroidetes, implying there was no significant change between GA and healthy control at the phylum level (Figure 2E and Supplementary Figure 2). In the second trimester, 10 bacterial taxa, including Megamonas, Veillonella, Paraprevotellaceae, Gemellales, Cetobacterium, Gemellaceae, Haemophilus, Pasteurellales, Pasteurellaceae, and Kluyvera were observed enriched in GA versus control (LDA > 2, FDR < 0.05, Supplementary Table 3). Megamonas, Veillonella, and Haemophilus were confirmed to show differential abundances between GA and healthy control after validated by MaAsLin analysis. On the contrary, Lachnospiraceae and Blautia were enriched in control (Figure 2F). Detailed information was shown in Supplementary Table 4. In the third trimester, after adjusting for covariates, increased abundance in Veillonella and decreased abundance in Lachnospiraceae and Blautia were observed in GA (Supplementary Figure 2).
Microbial Co-abundance Group Network
Since bacteria work as functional groups (Zhang et al., 2015), in the second trimester, the top 99 most abundant genera were clustered into 8 CAGs according to their co-abundance correlations (Figures 3A, B), and the regularized partial correlation network based on lasso regression was constructed (Figure 4). CAG1, CAG2, CAG3, and CAG4, accounting for 86.36% of ASVs, were consistently abundant both in GA and healthy control. Intriguingly, the significantly enriched taxa in GA belonged to CAG6 and the significantly enriched taxa in healthy control were specific to CAG2. These findings suggested a highly coordinated microbial regulatory network might underlie the occurrence of gestational anemia. Detailed information on CAG clusters was shown in Supplementary Table 5.
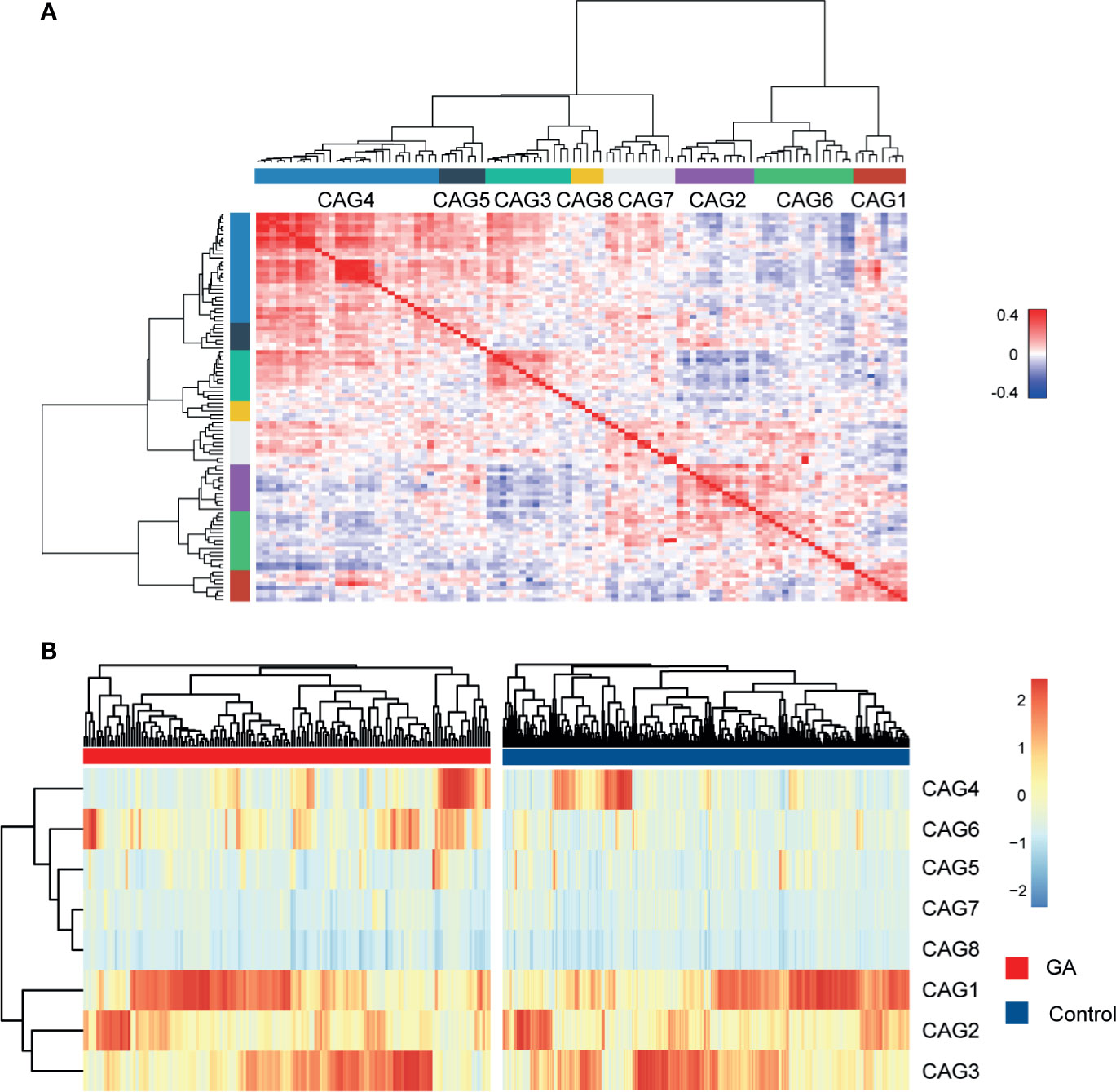
Figure 3 Heatmap of microbial co-abundance group. (A) Kendall correlations coefficients between the top 99 most abundant genera in the second trimester were calculated, and eight CAGs were clustered based on Kendall correlation matrix. (B) CAG differently enriched in GA and healthy control in the second trimester. GA, gestational anemia; CAG, co-abundance group.
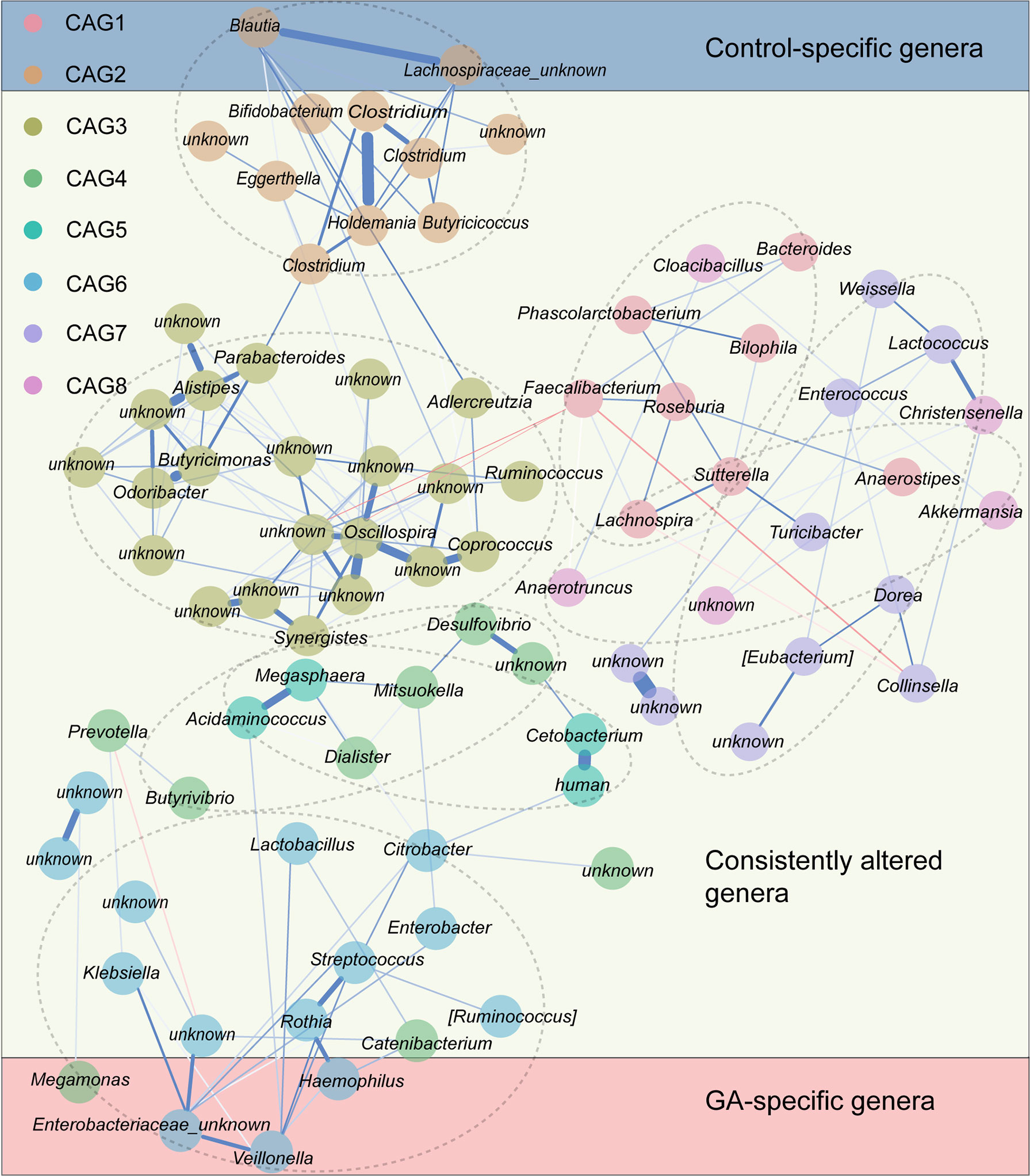
Figure 4 Co-abundance group network reflecting microbial changes in GA and healthy control. Regularized partial correlation network of top altered taxa in GA in the second trimester. Each node represented a taxon, and each edge represented the strength of partial correlation between two taxa. Edge weights represented the partial correlation coefficients. Blue edge represented positive correlation, and red edge represented negative correlation. GA, gestational anemia.
Interrelationship Between Gut Microbiota Composition, Clinical Indices, and GA
The abundance comparison adjusted for potential confounders showed decreased albumin (ALB), direct bilirubin (DB), free thyroxine (FT4), Hb, red blood cell (RBC), and total bilirubin (TB) were significantly associated with GA in the second trimester (Figure 5A and Supplementary Table 6). Intriguingly, no significant association was observed between GA and serum iron after adjusting for covariates (Figure 5B and Supplementary Table 7). Considering an FDR of 5%, the partial Spearman correlation delineated that 13 differentially abundant bacterial taxa were significantly correlated with clinical indices (Figure 5C and Supplementary Tables 8, 9). As the Sankey plots demonstrated, in the second trimester, Megamonas was negatively correlated with FT4 and further negatively correlated with GA, and Blautia was positively correlated with Hb and further negatively correlated with GA, indicating that gut microbiota could be involved in occurrence of anemia by interacting with clinical indices (Figure 5D).
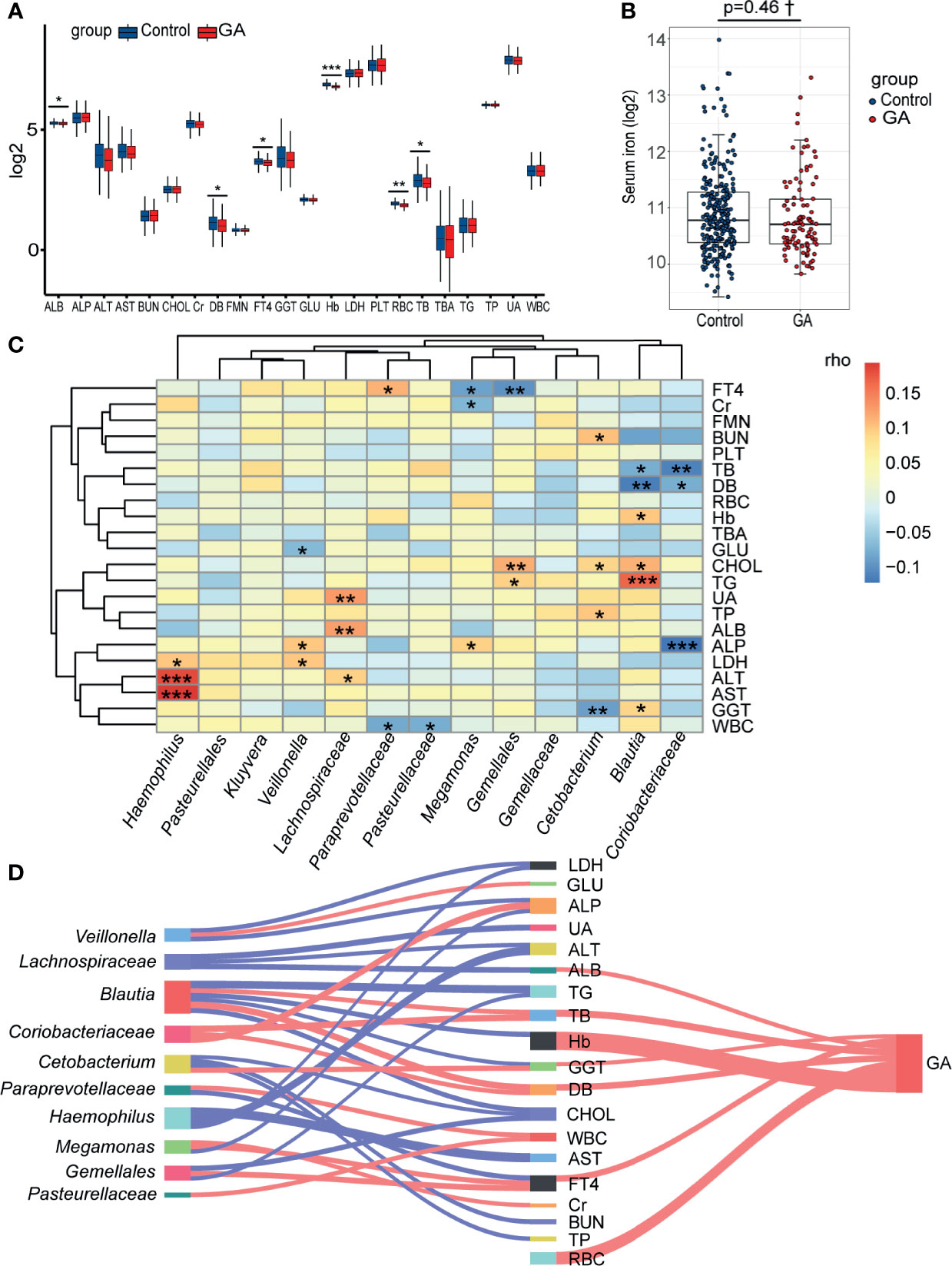
Figure 5 Correlations among the gut microbiota, clinical indices, and GA. (A) The box plot showed that the clinical indices significantly changed between two groups. (B) The difference of levels of serum iron in the second trimester between the two groups. (C) The heatmap of the partial Spearman correlations between gut microbiota and clinical indices in the second trimester (FDR < 0.05). (D) Relationships among gut microbiota composition, clinical indices, and GA (only significant correlations were presented, FDR < 0.05). The width of lines represented the partial correlation coefficients. Red line represented negative correlation and blue line represented positive correlation. GA, gestational anemia; FDR, false discovery rate; *P < 0.05, **P < 0.01, ***P < 0.001; †Adjusted for potential covariates.
Potential Predictive Value of Gut Microbial Signatures for GA
The cohort was further randomly divided into a Discovery set (GA: 91; Control: 244) and a Validation set (GA: 65; Control: 158). As shown in Figure 6, clinical indices alone had a poor performance in predicting upcoming anemia (Discovery AUC: 0.5112, 95%CI: 0.4421, 0.5802; Validation AUC: 0.5014, 95%CI: 0.4169, 0.5859). Further on, the potential value of gut microbiota acting as predictor was assessed. Using five genera adjusted by MaAsLin generated an AUC of 0.5701 (95%CI: 0.5033, 0.6368) in Discovery set and 0.5993 (95%CI: 0.5181, 0.6805) in Validation set. Using 13 genera based on LEfSe yielded an AUC of 0.6958 (95%CI: 0.6351, 0.7565) in Discovery set and 0.6820 (95%CI: 0.5939, 0.7702) in Validation set. Of note, CAG accurately predicted an upcoming GA with an AUC of 0.7738 (95%CI: 0.7171, 0.8306) and the classifying ability was further validated in Validation set (0.8223, 95%CI: 0.7573, 0.8874). No significant improved predictive performance was observed when including the combination of CAG and clinical indices into the prediction model (Discovery AUC: 0.7607, 95%CI: 0.7019, 0.8196; Validation AUC: 0.8382, 95%CI: 0.7770, 0.8994).
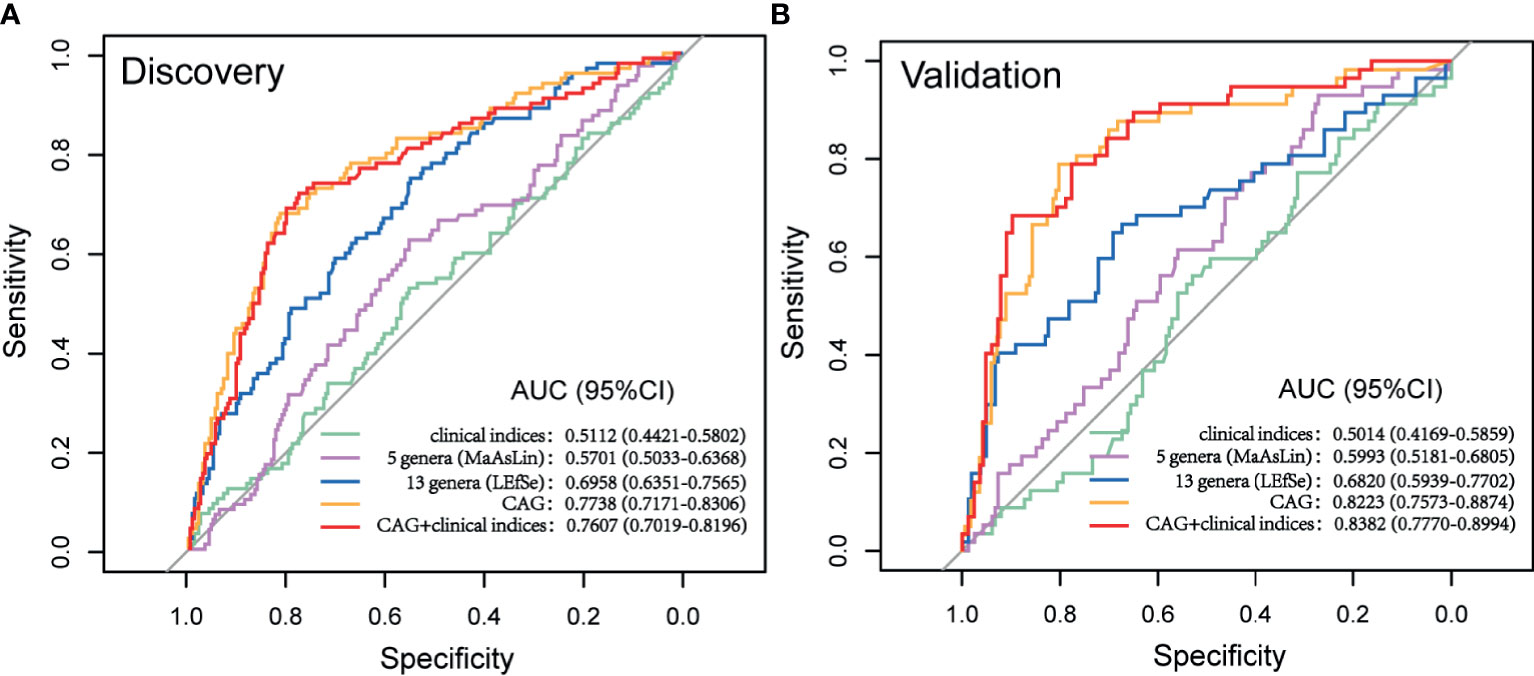
Figure 6 Prediction model of GA based on microbial signatures. (A) Discovery set (GA: 91; control: 244). (B) Validation set (GA: 65, control: 158). GA, gestational anemia; AUC, area under curve.
Discussion
In the current study, we delineated that GA microbial dysbiosis was characterized by several bacterial genera and structured CAG. A cross-sectional anemia classifier in the first trimester and second trimester has been constructed (Long et al., 2021); however, limited efforts have been made to prospectively predict future anemic women. The greatest advantage was that for the first time, a prediction model for upcoming anemia in normal pregnant women with relatively high discriminatory power was established based on novel gut microbial signatures.
It was challenging to determine the etiology of anemia and confirm IDA in pregnancy. Apart from Hb, more laboratory examinations (e.g., serum iron, transferrin receptor, transferrin saturation, and bone marrow iron) should be taken into consideration. The most common true anemia in pregnancy was IDA. In addition, there were no other types of pathological anemia (e.g., aplastic anemia and hemolytic anemia) remaining in our study. Thus, anemic women were basically postulated to suffer from IDA.
Clinical indices serve as generally accepted diagnostic criteria for GA cross-sectionally; however, they were confirmed to have very limited predictive value for potential impending anemia in normal pregnant women according to our study. Gut microbial signatures exhibited impressive performance in the prediction model. Bacteria were significantly differently abundant in GA and healthy control in the second trimester. Of note, 93.03% of healthy control and 97.44% of anemic women took folic acid and iron supplement from conception to the second trimester, implying the sufficient iron storage. From the perspective of potential mechanism, the altered gut microbiota in the early stage was conjectured to be subsequently associated with the altered health condition (i.e., from iron sufficiency to iron deficiency or malnutrition) and further accounted for the upcoming anemia.
Megamonas, Veillonella, and Haemophilus were enriched in GA. Megamonas could act as a beneficial bacterium, and it has been reported that compared to healthy microbiota, canine anal furunculosis (CAF) microbiota showed a decreased abundance of Megamonas (Maldonado-Contreras et al., 2020). Nevertheless, another study illustrated infant vitamin D supplementation was associated with a lower abundance of Megamonas in gut microbiota, implying the potential competitive relationship between vitamin D and Megamonas (Drall et al., 2020). Veillonella species, documented as the Fe (III)-reducing genera, were capable of supplying Fe (II) to combine with oxygen in Hb (Jin et al., 2019). We hypothesized there might be a negative feedback regulation, host generating more Veillonella species when detecting less Hb combined with Fe (II). Haemophilus is a genus of Gram-negative, containing several markedly pathogenic bacteria, such as Haemophilus influenzae causing septicemia. Indigenous bacteria might inhibit host Fe transport and storage via producing metabolites that suppress hypoxia-inducible factor 2α (HIF-2α), assumed as a master transcription factor of intestinal Fe absorption and increasing the Fe-storage protein ferritin (Das et al., 2020). Decreased incidence of Blautia has been detected in the gut microbiota of obese children and Blautia genera might help to reduce inflammation causally linked to obesity-related complications (Benítez-Páez et al., 2020).
Microbial network has been an increasingly popular tool to explore microbial community structure (Röttjers and Faust, 2018). Ecologically, gut microbiota exists in functional groups named “guilds” rather than isolation and thrives in communities with large numbers and develops close interactions, which are critical evolutionary pressures for natural selection in microbial evolution (Faust and Raes, 2012; Ma et al., 2020). We sought to reduce the dimensionality of microbial datasets to identify GA more effectively based on CAG, and interestingly, the prediction model exhibited a much higher discriminatory power.
It is noticeable that normal physiologic changes in pregnancy lead to a relative or absolute reduction in Hb concentration. However, it is still an open question that this “anemia” is physiologic or pathologic. Given gut microbial signatures were free from influence of hydremia, a better diagnostic or predictive performance using gut microbial signatures could be achieved when it comes to either true anemia or physiologic anemia.
There were some advantages of our study. Firstly, samples were collected prior to treatment initiation in a large and well-characterized cohort. Secondly, antibiotics use was controlled in analysis. On the other hand, most of pregnant women enrolled in the study claimed antibiotics were used very early in pregnancy at a low dose and once they were discovered to be pregnant, antibiotics use was avoided as much as possible, which meant a possible lesser impact of antibiotics on gut microbiota. Thirdly, we constituted the first exploration to prospectively predict the risk of anemia in healthy subjects based on gut microbiota. Lastly, a much more accurate prediction model was built based on CAG network.
There were several limitations. Firstly, as we have discussed above, diagnostic evidence of IDA was not sufficient enough. Secondly, we did not construct a GA classifier using gut microbiota in the third trimester since that Hb was supposed to be a better alternative, quicker and costing lower. There was no information on vitamin D levels or supplementation in women, which was supposed to be a factor associated to gut dysbiosis and consequently GA. In addition, lack of metagenomics sequencing limited data interpretation from the angle of species level and bacterial function. Finally, there was not an independent cohort to verify the prediction model; our study merely provided evidence of association rather than causality and further studies are supposed to be conducted to validate the association.
Conclusions
Our results showed that alterations in the gut microbial community were associated with anemia in pregnancy. Moreover, microbial signatures relatively accurately predicted the subsequent incidence of anemia in normal pregnant women. Our findings could provide new insights into understanding the role of gut microbiota in GA, identifying high-risk individuals, and modulating gut microbiota as a therapeutic target, thus improving quality of life and well-being of women and children.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics Statement
The studies involving human participants were reviewed and approved by the Ethics Committee of Nanjing Medical University [FWA00001501 No. (2017) 003]. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
Conceptualization: XW and YX. Formal analysis: HW and SD. Methodology: HW and YQ. Writing original draft: HW and SD. Verifying the underlying data: YQ and XY. Review and editing: YQ, XY, TC, XW, and YX. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by China-U.S. Program for Biomedical Collaborative Research (NSFC-NIH) (81961128022), the fifth phase of “333 High-level Talent Training Project” of the Jiangsu Province (BRA2020070), Postgraduate Research & Practice Innovation Program of Jiangsu Province (JX10313741), and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors would like to acknowledge and are grateful for all the patient participants, infants, and their families that made this study possible.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2021.734561/full#supplementary-material
Supplementary Figure 1 | Flow chart of the inclusion of subjects.
Supplementary Figure 2 | (A) Relative proportions of bacterial phyla in GA and healthy control in the third trimester. (B) Histogram of the LDA scores computed for differentially abundant taxa between GA and healthy control in the third trimester. GA, gestational anemia; LDA, linear discriminant analysis; *Genera remained significantly associated with GA after adjusting for covariates using multivariate association with linear models algorithm (MaAsLin).
References
Anker, S. D., Comin Colet, J., Filippatos, G., Willenheimer, R., Dickstein, K., Drexler, H., et al. (2009). Ferric Carboxymaltose in Patients With Heart Failure and Iron Deficiency. N. Engl. J. Med. 361 (25), 2436–2448. doi: 10.1056/NEJMoa0908355
Bäckhed, F., Ding, H., Wang, T., Hooper, L. V., Koh, G. Y., Nagy, A., et al. (2004). The Gut Microbiota as an Environmental Factor That Regulates Fat Storage. Proc. Natl. Acad. Sci. U.S.A. 101 (44), 15718–15723. doi: 10.1073/pnas.0407076101
Balamurugan, R., Mary, R. R., Chittaranjan, S., Jancy, H., Shobana Devi, R., Ramakrishna, B. S. (2010). Low Levels of Faecal Lactobacilli in Women With Iron-Deficiency Anaemia in South India. Br. J. Nutr. 104 (7), 931–934. doi: 10.1017/s0007114510001637
Benítez-Páez, A., Gómez Del Pugar, E. M., López-Almela, I., Moya-Pérez, Á., Codoñer-Franch, P., Sanz, Y. (2020). Depletion of Species in the Microbiota of Obese Children Relates to Intestinal Inflammation and Metabolic Phenotype Worsening. mSystems 5 (2), e00857–19. doi: 10.1128/mSystems.00857-19
Camaschella, C. (2015). Iron-Deficiency Anemia. N. Engl. J. Med. 372 (19), 1832–1843. doi: 10.1056/NEJMra1401038
Das, N. K., Schwartz, A. J., Barthel, G., Inohara, N., Liu, Q., Sankar, A., et al. (2020). Microbial Metabolite Signaling Is Required for Systemic Iron Homeostasis. Cell Metab. 31 (1), 115–130.e6. doi: 10.1016/j.cmet.2019.10.005
Drall, K. M., Field, C. J., Haqq, A. M., de Souza, R. J., Tun, H. M., Morales-Lizcano, N. P., et al. (2020). Vitamin D Supplementation in Pregnancy and Early Infancy in Relation to Gut Microbiota Composition and Colonization: Implications for Viral Respiratory Infections. Gut Microbes 12 (1), 1799734. doi: 10.1080/19490976.2020.1799734
Epskamp, S., Fried, E. I. (2018). A Tutorial on Regularized Partial Correlation Networks. Psychol. Methods 23 (4), 617–634. doi: 10.1037/met0000167
Faust, K., Raes, J. (2012). Microbial Interactions: From Networks to Models. Nat. Rev. Microbiol. 10 (8), 538–550. doi: 10.1038/nrmicro2832
Figueiredo, A., Gomes-Filho, I. S., Silva, R. B., Pereira, P. P. S., Mata, F., Lyrio, A. O., et al. (2018). Maternal Anemia and Low Birth Weight: A Systematic Review and Meta-Analysis. Nutrients 10 (5), 601. doi: 10.3390/nu10050601
Goonewardene, M., Shehata, M., Hamad, A. (2012). Anaemia in Pregnancy. Best Pract. Res. Clin. Obstet. Gynaecol. 26 (1), 3–24. doi: 10.1016/j.bpobgyn.2011.10.010
Guignard, J., Deneux-Tharaux, C., Seco, A., Beucher, G., Kayem, G., Bonnet, M. P. (2021). Gestational Anaemia and Severe Acute Maternal Morbidity: A Population-Based Study. Anaesthesia 76 (1), 61–71. doi: 10.1111/anae.15222
Human Microbiome Project Consortium. (2012). Structure, Function and Diversity of the Healthy Human Microbiome. Nature 486 (7402), 207–214. doi: 10.1038/nature11234
Hunt, J. M. (2002). Reversing Productivity Losses From Iron Deficiency: The Economic Case. J. Nutr. 132 (4 Suppl), 794s–801s. doi: 10.1093/jn/132.4.794S
Imhann, F., Vich Vila, A., Bonder, M. J., Fu, J., Gevers, D., Visschedijk, M. C., et al. (2018). Interplay of Host Genetics and Gut Microbiota Underlying the Onset and Clinical Presentation of Inflammatory Bowel Disease. Gut 67 (1), 108–119. doi: 10.1136/gutjnl-2016-312135
Jin, Z., Zhao, Z., Zhang, Y. (2019). Potential of Direct Interspecies Electron Transfer in Synergetic Enhancement of Methanogenesis and Sulfate Removal in an Up-Flow Anaerobic Sludge Blanket Reactor With Magnetite. Sci. Total Environ. 677, 299–306. doi: 10.1016/j.scitotenv.2019.04.372
Khan, K. S., Wojdyla, D., Say, L., Gülmezoglu, A. M., Van Look, P. F. (2006). WHO Analysis of Causes of Maternal Death: A Systematic Review. Lancet 367 (9516), 1066–1074. doi: 10.1016/s0140-6736(06)68397-9
Khosravi, A., Yáñez, A., Price, J. G., Chow, A., Merad, M., Goodridge, H. S., et al. (2014). Gut Microbiota Promote Hematopoiesis to Control Bacterial Infection. Cell Host Microbe 15 (3), 374–381. doi: 10.1016/j.chom.2014.02.006
Koren, O., Goodrich, J. K., Cullender, T. C., Spor, A., Laitinen, K., Bäckhed, H. K., et al. (2012). Host Remodeling of the Gut Microbiome and Metabolic Changes During Pregnancy. Cell 150 (3), 470–480. doi: 10.1016/j.cell.2012.07.008
Liang, L., Rasmussen, M. H., Piening, B., Shen, X., Chen, S., Röst, H., et al. (2020). Metabolic Dynamics and Prediction of Gestational Age and Time to Delivery in Pregnant Women. Cell 181 (7), 1680–1692.e1615. doi: 10.1016/j.cell.2020.05.002
Liu, H., Chen, X., Hu, X., Niu, H., Tian, R., Wang, H., et al. (2019). Alterations in the Gut Microbiome and Metabolism With Coronary Artery Disease Severity. Microbiome 7 (1), 68. doi: 10.1186/s40168-019-0683-9
Li, F., Zhang, C., Chen, Y. G., Shen, J. Y., Jiang, S., Li, J., et al. (2018). [Analysis on Maternal Anemia Rate and Related Factors in Taicang of Jiangsu Province in 2014-2016]. Zhonghua Yu Fang Yi Xue Za Zhi 52 (7), 703–708. doi: 10.3760/cma.j.issn.0253-9624.2018.07.005
Long, Y., Liang, F., Guo, R., Zhu, C., Zhao, X., Wang, X., et al. (2021). Gut Microbiota Signatures in Gestational Anemia. Front. Cell Infect. Microbiol. 11:549678. doi: 10.3389/fcimb.2021.549678
Lund, C. J., Donovan, J. C. (1967). Blood Volume During Pregnancy. Significance of Plasma and Red Cell Volumes. Am. J. Obstet Gynecol 98 (3), 394–403. doi: 10.1016/0002-9378(67)90160-3
Maldonado-Contreras, A., Ferrer, L., Cawley, C., Crain, S., Bhattarai, S., Toscano, J., et al. (2020). Dysbiosis in a Canine Model of Human Fistulizing Crohn's Disease. Gut Microbes 12 (1), 1785246. doi: 10.1080/19490976.2020.1785246
Ma, B., Wang, Y., Ye, S., Liu, S., Stirling, E., Gilbert, J. A., et al. (2020). Earth Microbial Co-Occurrence Network Reveals Interconnection Pattern Across Microbiomes. Microbiome 8 (1), 82. doi: 10.1186/s40168-020-00857-2
Morgan, X. C., Tickle, T. L., Sokol, H., Gevers, D., Devaney, K. L., Ward, D. V., et al. (2012). Dysfunction of the Intestinal Microbiome in Inflammatory Bowel Disease and Treatment. Genome Biol. 13 (9), R79. doi: 10.1186/gb-2012-13-9-r79
Prema, K., Ramalakshmi, B. A., Madhavapeddi, R., Babu, S. (1982). Immune Status of Anaemic Pregnant Women. Br. J. Obstet. Gynaecol. 89 (3), 222–225. doi: 10.1111/j.1471-0528.1982.tb03619.x
Rahman, M. M., Abe, S. K., Rahman, M. S., Kanda, M., Narita, S., Bilano, V., et al. (2016). Maternal Anemia and Risk of Adverse Birth and Health Outcomes in Low- and Middle-Income Countries: Systematic Review and Meta-Analysis. Am. J. Clin. Nutr. 103 (2), 495–504. doi: 10.3945/ajcn.115.107896
Röttjers, L., Faust, K. (2018). From Hairballs to Hypotheses-Biological Insights From Microbial Networks. FEMS Microbiol. Rev. 42 (6), 761–780. doi: 10.1093/femsre/fuy030
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic Biomarker Discovery and Explanation. Genome Biol. 12 (6), R60. doi: 10.1186/gb-2011-12-6-r60
Sifakis, S., Pharmakides, G. (2000). Anemia in Pregnancy. Ann. N Y Acad. Sci. 900, 125–136. doi: 10.1111/j.1749-6632.2000.tb06223.x
Thomas, A. M., Manghi, P., Asnicar, F., Pasolli, E., Armanini, F., Zolfo, M., et al. (2019). Metagenomic Analysis of Colorectal Cancer Datasets Identifies Cross-Cohort Microbial Diagnostic Signatures and a Link With Choline Degradation. Nat. Med. 25 (4), 667–678. doi: 10.1038/s41591-019-0405-7
Vich Vila, A., Imhann, F., Collij, V., Jankipersadsing, S. A., Gurry, T., Mujagic, Z., et al. (2018). Gut Microbiota Composition and Functional Changes in Inflammatory Bowel Disease and Irritable Bowel Syndrome. Sci. Transl. Med. 10 (472), eaap8914. doi: 10.1126/scitranslmed.aap8914
Wei, Y., Li, Y., Yan, L., Sun, C., Miao, Q., Wang, Q., et al. (2020). Alterations of Gut Microbiome in Autoimmune Hepatitis. Gut 69 (3), 569–577. doi: 10.1136/gutjnl-2018-317836
Wirbel, J., Pyl, P. T., Kartal, E., Zych, K., Kashani, A., Milanese, A., et al. (2019). Meta-Analysis of Fecal Metagenomes Reveals Global Microbial Signatures That are Specific for Colorectal Cancer. Nat. Med. 25 (4), 679–689. doi: 10.1038/s41591-019-0406-6
Zhang, Y., Chen, T., Zhang, Y., Hu, Q., Wang, X., Chang, H., et al. (2021). Contribution of Trace Element Exposure to Gestational Diabetes Mellitus Through Disturbing the Gut Microbiome. Environ. Int. 153, 106520. doi: 10.1016/j.envint.2021.106520
Zhang, C., Yin, A., Li, H., Wang, R., Wu, G., Shen, J., et al. (2015). Dietary Modulation of Gut Microbiota Contributes to Alleviation of Both Genetic and Simple Obesity in Children. EBioMedicine 2 (8), 968–984. doi: 10.1016/j.ebiom.2015.07.007
Keywords: 16S rRNA gene sequencing, gut microbiota, co-abundance group, prediction, the risk of developing gestational anemia
Citation: Wei H, Deng S, Qin Y, Yang X, Chen T, Wang X and Xia Y (2021) Insight Into the Potential Value of Gut Microbial Signatures for Prediction of Gestational Anemia. Front. Cell. Infect. Microbiol. 11:734561. doi: 10.3389/fcimb.2021.734561
Received: 01 July 2021; Accepted: 09 August 2021;
Published: 30 August 2021.
Edited by:
Gislane Lelis Vilela de Oliveira, São Paulo State University, BrazilReviewed by:
Huajun Zheng, Shanghai Institute for Biomedical and Pharmaceutical Technologies, ChinaFrancesco Strati, European Institute of Oncology (IEO), Italy
Júlia Teixeira Cottas De Azevedo, Hemocentro Foundation of Ribeirão Preto, Brazil
Copyright © 2021 Wei, Deng, Qin, Yang, Chen, Wang and Xia. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yankai Xia, yankaixia@njmu.edu.cn; Xu Wang, sepnine@njmu.edu.cn
†These authors have contributed equally to this work and share first authorship
 Hongcheng Wei1,2†
Hongcheng Wei1,2†  Siting Deng
Siting Deng Yufeng Qin
Yufeng Qin Xu Wang
Xu Wang Yankai Xia
Yankai Xia