Abstract
Bacillus species genomes are rich in plant growth-promoting genetic elements. Bacillus subtilis and Bacillus velezensis are important plant growth promoters; hence, to further improve their abilities, the genetic elements responsible for these traits were characterized and reported. Genetic elements reported include those of auxin, nitrogen fixation, siderophore production, iron acquisition, volatile organic compounds, and antibiotics. Furthermore, the presence of phages and antibiotic-resistant genes in the genomes are reported. Pan-genome analysis was conducted using ten Bacillus species. From the analysis, pan-genome of Bacillus subtilis and Bacillus velezensis are still open. Ultimately, this study brings an insight into the genetic components of the plant growth-promoting abilities of these strains and shows their potential biotechnological applications in agriculture and other relevant sectors.
Similar content being viewed by others
Introduction
The challenges of climate change and urbanization impact on food production has necessitated the need to bring up solutions that will mitigate these effects. The health hazard posed by chemical fertilizers is a cause for concern; hence, the need for an environmental and health-friendly approach. This led to the use of microorganisms for food production. These microorganisms, termed plant growth-promoting bacteria (PGPB), increase food production through direct and indirect means such as phytohormone production, nitrogen fixation, cyanide production, siderophore production, antibiotic production, and phosphorous solubilization. These mechanisms have been reviewed in various studies [25, 31, 39, 43, 47]. They produce antifungal and antibacterial compounds to help plants against pathogens. Furthermore, they act in the bioremediation of contaminated soil. Contaminations in the form of salinity and heavy metal pollution are taken care of by PGPB. Several genus of bacteria support plant growth promotions; however, this study will focus on the Bacillus genus.
Bacillus genus is abundant in the rhizobiome. They are present in the rhizosphere of several crop plants where they support plant growth. They are good root colonizers and produce several metabolites, which makes them good biocontrol agents against plant pathogens. Reports on their biocontrol activities have been studied by various groups of researchers [3, 15, 21, 46, 50]. The most important feature of Bacillus spp. is their ability to form spores. Spores help to protect them against various stress conditions, thus, enabling their application for plant growth promotion, bioremediation, and industrial applications.
Furthermore, comparative microbial genomics based on sequence similarity will help in identifying the important genetic contents shared among all plant growth promoting isolates as well as subset of genes encoding biocontrol and novel functions. Pan-genome represents species genomic diversity and it includes both the core and variable genome content [11]. Pan-genome aids in taxonomic classifications (phylogenomic analysis), precise determination of genomic contents of a group (calculation of core, pan, and variable genome), and organism’s lifestyle (allopatric or sympatric) [13].
This study focuses on three strains of this genus, viz. Bacillus subtilis A1 (BSA1), Bacillus velezensis A3 (BVA3), and Bacillus subtilis A29 (BSA29). In a previous study [38], these strains promote the growth of maize plants on the field. In vitro assay showed their efficiency in inhibiting Fusarium graminearum. Furthermore, they were able to solubilize phosphate, produce IAA, siderophore, protease, oxidase, and ACC deaminase enzyme, among others. In another study [36], they produce several volatile organic compounds (VOCs) that are related to their biocontrol ability. In the present study, therefore, we report the genomic characterization and the genetic basis of the plant growth-promoting traits exhibited by these strains. The presence of phages in these strains was also reported in the study. We have used comparative genomics approach to unravel the plant growth promoting and biocontrol potential of the strains in this study. The result obtained can be applied for the genetic modification of these strains for various plant growth-promoting abilities.
Materials and Methods
Isolation, Identification, Antimicrobial Activity, and Plant Growth Promotion Assay
Isolation of bacterial strains and identification, as well as plant growth promotion assay, are reported in a previous study [38]. The isolates are Bacillus subtilis A1, Bacillus velezensis A3, and Bacillus subtilis A29. In brief, 31 isolates were screened for their plant growth-promoting traits, and 3 were selected for field trials [38]. They showed improvement in maize growth compared to the control when inoculated in single and consortia.
Genome Sequencing and Annotation
The genomic DNA was extracted from overnight cultures in LB medium [12] using a ZR soil microbe DNA MiniPrep extraction kit (Zymo Research, USA), following the manufacturer’s instructions. The DNA quality and quantity were determined using a NanoDrop Lite spectrophotometer (Thermo Fisher Scientific, CA, USA). The genomes of the strains were sequenced on an Illumina HiSeq sequencer at Molecular Research (MR DNA), Shallowater, TX. The libraries were prepared using Kapa HyperPlus kits (Roche), following the manufacturer’s user guide. The initial concentration of DNA was evaluated using the Qubit double-stranded DNA (dsDNA) high-sensitivity (HS) assay kit (Life Technologies). A total of 25 ng of DNA were used to prepare the libraries. The protocol starts with enzymatic fragmentation to produce dsDNA fragments, followed by end repair and A-tailing to produce end-repaired 5′-phosphorylated 3′-deoxyribosyladenine (dA)-tailed dsDNA fragments. In the adapter ligation step, dsDNA adapters are ligated to 3′-dA-tailed molecules. The final step is library amplification, which employs high-fidelity, low-bias PCR to amplify library fragments carrying appropriate adapter sequences on both ends. Following the library preparation, the final concentrations of the libraries were measured using the Qubit dsDNA HS assay kit (Life Technologies), and the average library size was determined using the Agilent 2100 Bioanalyzer (Agilent Technologies). Bacillus subtilis A1, Bacillus velezensis strain A3, and Bacillus subtilis strain A29 DNA concentrations are 114.0, 84.8, and 187.0 ng/μl, respectively, while the final library DNA concentrations are 62.0, 62.0, and 58.8 ng/μl, respectively. The average library sizes of Bacillus subtilis A1, Bacillus velezensis strain A3, and Bacillus subtilis strain A29 are 680, 694, and 695 bp, respectively. The libraries were pooled, diluted (to 9.0 pM), and paired-end sequenced for 500 cycles using the HiSeq system (Illumina), with an average read length of 2 × 250 bp.
The raw sequences were processed to obtain high-quality reads using the Kbase [20] platform. The quality of the reads was checked using FastQC (v.1.0.4) [6]. The reads were trimmed to remove adapters and low-quality sequences using Trimmomatic (v.0.36) [16], with the default parameters. The reads were assembled by de novo assembly using SPAdes v.3.12.0 [35], with the default parameters. Gene function prediction was performed using the Rapid Annotations using Subsystems Technology (RAST v.2.0) server (http://rast.nmpdr.org) [8].
The Genbank accession numbers are SHOB00000000, SHOC00000000, and SHOD00000000, while the BioProject accession numbers are PRJNA516328, PRJNA516332, and PRJNA516331, respectively. The Sequence Read Archive (SRA) has accession numbers SRR8540661, SRR8550002, and SRR8541016, respectively.
Genome Mining for BGCs, Antibiotic-Resistant Genes, Virulent Factors, and Phages
The genome sequences of the selected strains were determined and mining for biosynthetic gene clusters of antimicrobial compounds, including NRPs, PKs, NRPs-PKs hybrids, bacteriocins, and terpenes, was conducted with RAST system [8, 17, 40], antiSMASH 5.0 [14], and BAGEL4 [48] using the default settings. Each draft genome was assembled into a pseudomolecule using a closely related strain as a reference before applying to the pipelines. Antimicrobial resistance genes were mined using the Resistance Gene Identifier (RGI) tool of the Comprehensive Antibiotic Resistance Database (CARD)4 [4] using contigs file with the parameters “Perfect and strict hits only” and “High quality/coverage.” The presence of phages was checked using phaster [7] with default settings.
Pan-genome and Core-genome Analyses
Bacterial pan-genome analysis (BPGA) is a high-speed and highly efficient computational pipeline used for comparative genomic analysis and pan-genome construction [19]. Pan-genome and core-genome of 10 Bacillus species (Table 3) were obtained by BPGA pipeline. Power-law regression based on Heaps’ law was used to calculate curve fitting of the pan-genome as follows:
y ¼ A pan x^ðB panÞ þ C pan.
where y is the pan-genome size, x is the genome number, and A_pan, B_pan, and C_pan are fit parameters. B_pan is equivalent to the parameter c and a = 1 − c. According to Heaps’ law, a pan-genome is open when 0\c\1 and a B 1 and close when c\1 and a[1. The exponential regression model y ¼ A core e^ðB core _ xÞ þ C core was used to calculate curve fitting of the core-genome, where y is the core-genome size, x is the genome number, e is the Euler number, and A_core, B_core, and C_core are fit parameters [45].
USEARCH clustering algorithm with a 50% cut-off was utilized for clustering genes. Pan- and core-genome plot was generated using the default settings while MUSCLE was used for aligning genes and phylogeny was analyzed with the neighbor-joining method.
COG and KEGG Functional Classification of Genes
Downstream analysis of data subsets under KEGG/COG categories deciphers the BPGA platform for further “omics” approaches. BPGA employs the ublast function of USEARCH to identify best hits with respective reference databases and classify them according to KEGG and COG categories [19].
Phylogenetic Analysis
Phylogenetic tree was constructed based on the average nucleotide identity (ANI). The overall similarity between the whole-genome sequences was calculated using the Orthologous Average Nucleotide Identity Tool (OAT) v0.93.1 [54].
Results and Discussion
General Characterization of the Strains
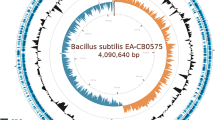
The genome map of the three isolates presented in Fig. 1 shows the various genetic components of the isolates. Figure 2 shows the distribution of the gene categories in the isolates. In all three, gene for amino acids is more abundant followed by carbohydrate and protein metabolism.
Genetic Elements Involved in Stress Resistance, Soil, and Plant Colonization Abilities
Stress Resistance and Tolerance
Genome analysis of BSA1, BVA3, and BSA29 showed the presence of stress tolerance proteins in the genomes of these isolates. Osmotic stress resistance genes of the “Opu” family are present in the three genomes (Table S1). Enzymes such as betaine aldehyde dehydrogenase and alcohol dehydrogenase that help against osmotic stress are found in the genomes. One nitrosative stress resistance gene nsrR is also present in all the genomes. Superoxide dismutase enzyme, which is important in militating against oxidative stress, is present in the three genomes along with perR and fuR genes. Apart from the specific stress response proteins that have been mentioned, several general stress response proteins are also present in the genomes of these microbes. These include rsbS, rsbT, rsbW, rsbU, rsbR, rsbV, and hfq genes. The presence of stress response genes confirms the ability of BSA1, BSA29, and BVA3 to help plants in stress tolerance [1, 50]. Proteins involved in heat and cold stress are important in DNA and RNA stabilization, thereby increasing transcription and translation efficiency during the stress period [30].
Spore Formation
Several genes and enzymes regulating spore formation and dormancy are largely represented in the genomes of the three microbes (Table S1). Among these are the ypeB, ydhD, kapB, mazG, tasA, yqxM genes etc. while enzymes present include sporulation kinase B and sporulation kinase C. Several spore formation gene clusters are represented in these genomes. Bacillus species are known for spore formation. Spore production enhances their ability to cope with environmental stress such as drought and salinity [56].
Heavy Metal Resistance
The genomes show the presence of copper resistance genes, viz. copD and copC genes. Proteins regulating the transportation of metals, such as the copper-translocating P-type ATPase (EC 3.6.3.4) and the cobalt-zinc-cadmium resistance protein, are present in the genomes (Table S1). The presence of heavy metal-resistant genes shows that these strains can adapt favorably when exposed to heavy metals. Heavy metals deplete bacterial populations in the soil [27], hence reducing microbial-plant growth promotion efficacy. Therefore, microbial resistance to heavy metals is critical for their survival in the soil. These strains, having several heavy metal resistance genes, stand a chance to survive in heavy metal polluted environment.
Motility, Chemotaxis, and Attachment to Plant Surfaces
All forms of movement in Bacillus spp. are with the use of flagella. The flagellum is their medium of transport for food and attachment to host plants. Therefore, it is an important feature in these bacteria genera. Based on this, the genomes of these isolates revealed the presence of several flagella biosynthesis proteins and regulators such as flgB, flgD, and flgK. Cell division protein Fst1, which codes for the enzyme peptidoglycan synthetase (EC 2.4.1.129), is also present in the genomes (Table S1). The ability to move and attach to plant surfaces is important for efficient root colonization. Teichoic acid is fundamental in root colonization [53]. The genomes of these strains revealed the presence of genes involved in the production of teichoic acid.
Genetic Elements Involved in Plant Growth Promotion Activities
Biocontrol Activities and Antibiotic Resistance
Genomic analysis revealed the presence of several gene clusters involved in the production of antimicrobial compounds, including genes involved in bacteriocins, terpenes, PKS, and NRPs gene clusters (Tables 1 and S2, Figs. 3 and 4). Gene clusters involved in bacillibactin, fengycin, macrolactin H, subtilosin A, sporulation killing factor, and surfactin were reported (Tables 1 and S2, Figs. 3, 4, S1–S9). In addition, antibiotic resistance genes were detected in the genomes (Table 2, Figs. S10-S13). Genes coding for daptomycin resistance and tunicamycin resistance proteins, including the ykkC, ykkd, aadK, vmlR, and tmrB genes (Table 2, Figs. S10-S13); genes encoding for streptothricin resistance and Fosfomycin resistance protein (fosB); and genes coding for resistance to fluoroquinolones are present (Table S1). Bacillus strains, especially Bacillus subtilis, are good biocontrol agents [15, 21, 22].
The ability to produce toxic compounds against pathogens is important for the biological control of plant pathogens. Bacillus produce compounds such as fengycin, surfactin, bacilysin, bacillomycin, and subtilosin [2, 24]. Fengycin acts by inhibiting the growth of filamentous fungi [33]. The surfactin lipopeptides have antiviral activity and show some antifungal activity. Fengycin, bacillomycin, and surfactin act stronger in cooperation as biocontrol agents and in biofilm formation. This was concluded in the study of Zeriouh et al. [55] where they reported reduced production of biofilm due to the absence of surfactin. So much importance is attached to the antimicrobial production by Bacillus species in biocontrol. Bacillus mutants that were unable to produce surfactin, bacillomycin, and fengycin lost their ability to control various plant diseases [9].
Antimicrobial resistance is becoming a challenge to clinical, industrial, and agricultural sectors [26]. The presence of biofilm in Bacillus subtilis, especially, makes it a classic example in this challenge. Biofilms protect the microbes from being destroyed by antibiotics. To the plants, this helps in protecting the plant’s “fighters” from being destroyed by toxin-producing pathogens. The results in this study support and confirm that Bacillus subtilis [18] and Bacillus velezensis [32, 46] produce antifungal and antibacterial compounds for improved plant health.
Iron Acquisition
The three genomes contain genes involved in iron metabolism as well as siderophore biosynthesis, especially bacillibactin and anthrachelin class siderophores. Iron acquisition gene yycJ/Wa1J and heme transporter protein htsABC (Table S1) are present in the genomes of the species. Iron transport peroxidase efeB and permease efeU are present in the three genomes. Additionally, gene clusters involved in iron cluster assembly were found, and it contains paaD-like protein (DUF59), iscR, sufE2, sufD, sufC, sufB, and apbC genes (Table S1). Iron is important in the synthesis of chlorophyll, and it is important in the maintenance of chloroplast structure. Therefore, the availability of iron is crucial for plant’s survival. Siderophores are molecules produced by bacteria, which makes iron available for plant use [25]. Siderophore-producing bacteria improve iron availability for plant development [37]. The presence of siderophore-producing genes in the genomes of these strains shows that they will be able to sequester available iron in the rhizosphere for plant use.
Nitrogen, Sulfur, and Phosphorus Acquisition
Genes regulating the metabolism of nitrogen, sulfur, and phosphorous are present in the genomes of the three bacteria. Nitrite reductase, glutamine synthetase, and glutamate synthase genes are all present (Table S1). Nitrogen regulatory protein P-II class and ammonia transport proteins are present. Nitric oxide reductase genes norD and norQ are present. This shows the ability of these bacteria to counter nitrosative stress. Additionally, genes involved in sulfate metabolism were detected. Genes for disulfide reductase (Tpx and Bcp types) in all three genomes and genes for galactosylceramide and sulfatide metabolism (arylsulfatase gene) were found in the genomes of BSA1 and BSA29. Furthermore, phosphate metabolism was supported by the presence of manganese-dependent inorganic pyrophosphatase, alkaline phosphatase genes (phoP, phoH, and phoR), and pyrophosphatse gene ppax (Table S1). The major sources of nitrogen for plant are chemical fertilizers and biological nitrogen fixation [5] and nitrogen is the most limiting element in plant growth, hence the importance of the nitrogen-fixing ability in these strains. Coupled with the presence of sulfur and phosphate synthesis genes, these strains are capable of nitrogen-fixation and phosphate solubilization [23, 38]. The availability of these nutrients is essential and critical in plant growth promotion.
IAA Biosynthesis and Ɣ-Aminobutyric Acid (GABA) Metabolism
The three genomes reveal the presence of genes regulating auxin biosynthesis. These are tryptophan synthase, anthranilate phosphoribosyltransferase, and phosphoribosylanthranilate isomerase (Table S1). The genomes also possess gabR gene encoding GABA aminotransferase (Table S1). Phytohormones regulate plant growth and tolerance. Auxin, a major phytohormone, is essential for growth regulation and stress adaptation responses. GABA, on the other hand, is involved in signaling between rhizosphere microorganisms and plants [37]. The presence of IAA and GABA genes in the genomes of these isolates infers that they will be effective in mitigating plant stress while promoting plant growth and development.
Polyamine Production and Modulation of Ethylene Levels
Several polyamine biosynthesis and transport genes are found in the genomes of the Bacillus spp, including arcD gene encoding for arginine/ornithine antiporter, and genes encoding for enzymes involved in spermidine biosynthesis, which include spermidine synthase. Agmatinase involved in putrescine biosynthesis is also present in the genomes (Table S1). Polyamines are important in the plat growth promoting abilities of Bacillus strains. Xie et al. [51] reported in their study that spermidine production by Bacillus subtilis was found to inhibit the production of ethylene, which affects interactions between plants and microorganisms.
Plant Growth Promotion Activities by Modulation of VOCs
Biosynthesis of VOCs originates from sulfur, nitrogen, ketones, alcohols, and aldehyde compounds. The genome analysis of these species shows a high level of genes and enzymes involved in the biosynthesis of these compounds. Therefore, these isolates can produce a high number of VOCs. Besides, genes involved in the biosynthesis of acetoin and butanediol are present (Table S1). VOCs impact plant growth by acting as signal molecules [37] and as biocontrol agents. Bacillus spp. are known to produce diverse kinds of VOCs, which result from the compounds stated above. Genes and enzymes involved in the pathways of these compounds are present in the genomes of BSA1, BVA3, and BSA29 (Table S1). Genes and pathways involved in the metabolism of acetoin and butanediol are revealed by the genomic analysis. Bacillus subtilis and Bacillus velezensis are reported to be good producers of acetoin and butanediol, which are used in biocontrol activities [34, 41, 52].
Phage Synthesis
The genomes of the Bacillus spp. were investigated for the presence of prophages, plasmids, and insertion elements. The results showed at least one intact phage region for all strains (Fig. 5). Blast hits against the virus, and bacterial databases are shown in Table S3. No plasmid was found in any of the strains. Different regions and the number of coding sequences are shown in Table S3. These strains can be effectively engineered in developing phage therapies for pathogen biocontrol. Phages have been reported as good biocontrol agents against human, animal, plant, and foodborne pathogens [28, 42, 49].
Pan-genome Analysis
Bacterial genomes harbor core and accessory genes, which may be specific to an individual species. Core-genomes are present in all strains studied while the accessory genomes differentiate one specie from another [45]. The accessory genomes usually confer genes regulating species-specific advantageous traits such as metabolite production, antibiotic resistance, virulence mechanisms, plant growth and disease suppression, siderophore production, and/or growth hormone production.
In this study, we selected 10 Bacillus species (5 B. subtilis and 5 B. velezensis) based on their host (soil or plant) to estimate the pan- and core-genome sizes (Table S4). From our analysis, we conclude that the B. subtilis and B. velezensis have an open genome since the core/pan-genome ratio did not reach a distinct plateau (Fig. 6). However, the addition of more genomes might add to the number of accessory and unique genes, which is in line with the previous hypothesis by [10]. The pan-genome consisted of 777 core genes while BSA29 has the lowest number of accessory genes and unique genes, and Bacillus subtilis R31 has the highest number of accessory genes and unique genes (Table 3).
Phylogenomics
Our phylogenetic trees were based on analyses of the core-genome, and ANI of the 10 genomes used (Figs. 7 and 8). The findings further refine the relationships within the genus. The phylogenetic tree based on the core-genomes (Fig. 7a) revealed that BVA3 is closer to B. subtilis R31 than the other B. velezensis species while BSA1 and BSA29 are very much close to each other same as the pan phylogeny (Fig. 7b). However, the BVA3 position in the pan phylogeny is closer to BSA1 than the other B. velezensis species. Therefore, we can infer that BVA3 have the same ancestor with BSA1 and BSA29.
Despite the drawback that the resolution is normally not sufficient to fully separate sub-species and the fact that that it is also liable to biases due to primer sequence matching [44], 16S rRNA sequencing is the most used parameter to explore bacteria phylogenetic relationships. Phenotypical and biochemical traits have also been used; however, these traits can be affected by choice of culture medium and other conditions. Therefore, there is need for objective methods that show high resolution. One promising method is the use of average nucleotide identity which was described by Han et al. [29]. In this study, the OrthoANI analysis showed 100% average nucleotide identity between our isolates BSA1 and BSA29 while BVA3 shows 98.76% similarity with B. velezensis YB-130. When comparing all isolates from this study, values were between 98.34 and 100% (for B. subtilis species) and between 97.66 and 98.97% for the B. velezensis strains. The comparison between the B. subtilis strains and B. velezensis strains showed always identity values below 80%.
Functional Genome Analyses
To assign biological functions to the orthologs, the corresponding amino acids were annotated using COG. According to the COG analysis major category distribution, majority of the core proteins are for metabolism (40.3%), followed by information storage and processing (25.6%), 23.03% were poorly characterized while the remaining 11.01% are for cellular signaling and processing (Fig. S14a). In detail, the gene functions are as follows: cell cycle control, cell division, chromosome partitioning (2.43%), cell wall/membrane/envelope biogenesis (13.21%), cell motility(5.66%), post-translational modification, protein turnover, and chaperones (9.31%), signal transduction mechanisms (15.15%), intracellular trafficking, secretion, and vesicular transport (3.75%), defense mechanisms (10.9%), translation, ribosomal structure and biogenesis (10%), and transcription (30.64%). Others include those involved in replication, recombination and repair (21%), energy production and conversion (11.97%), carbohydrate transport and metabolism (19.02%), amino acid transport and metabolism (25.02%), nucleotide transport and metabolism (4.48%), coenzyme transport and metabolism (10.73%), lipid transport and metabolism (10.94%), secondary metabolites biosynthesis, catabolism (12.48%), inorganic ion transport and metabolism (13.74%), general function prediction only (39.04%), and genes with unknown functions (30.46%) (Fig. 10b). We also use KEGG to map cellular functions and the genes were divided according to the biological pathways they are likely to participate in (Fig. S15).
Conclusion
Bacillus species have great potentials in agriculture and biotechnology. They are good producers of biocontrol agents and plant growth-promoting molecules hence their various applications for plant growth promotion and biocontrol abilities on various crops. The genomic analysis of B. subtilis A1, B. velezensis A3, and B. subtilis A29 showed the presence of genes, enzymes, and pathways involved in many plant growth-promoting activities such as growth hormone production, VOCs production, siderophore production, nitrogen, phosphorous, and sulfur metabolism. Their capability to be used in developing biocontrol phages was also established in the presence of many phage regions in their genomes. The pan-genome of Bacillus subtilis and Bacillus velezensis are still open. Hence, these isolates are promising plant growth promoters and can improve food security.
References
Abd El-Daim, I. A., Bejai, S., & Meijer, J. (2019). Bacillus velezensis 5113 induced metabolic and molecular reprogramming during abiotic stress tolerance in wheat. Scientific Reports, 9, 1–18.
Adeniji, A. A., Aremu, O. S., & Babalola, O. O. (2019). Selecting lipopeptide-producing, Fusarium-suppressing Bacillus spp.: Metabolomic and genomic probing of Bacillus velezensis NWUMFkBS10.5. Microbiology Open, 8, e00742.
Ajilogba, C. F., & Babalola, O. O. (2019). GC–MS analysis of volatile organic compounds from Bambara groundnut rhizobacteria and their antibacterial properties. World Journal of Microbiology and Biotechnology, 35, 83.
Alcock, B. P., Raphenya, A. R., Lau, T. T. Y., Tsang, K. K., Bouchard, M., Edalatmand, A., Huynh, W., Nguyen, A.-L.V., Cheng, A. A., Liu, S., Min, S. Y., Miroshnichenko, A., Tran, H.-K., Werfalli, R. E., Nasir, J. A., Oloni, M., Speicher, D. J., Florescu, A., Singh, B., … McArthur, A. G. (2019). CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Research, 48, D517–D525.
Alori, E. T., Dare, M. O., & Babalola, O. O. (2017). Microbial inoculants for soil quality and plant health. Sustainable Agriculture Reviews, 281–307 https://doi.org/10.1007/978-3-319-48006-0_9
Andrews, S. (2010). FastQC: A quality control tool for high throughput sequence data. https://www.bioinformatics.babraham.ac.uk/projects/fastqc
Arndt, D., Grant, J. R., Marcu, A., Sajed, T., Pon, A., Liang, Y., & Wishart, D. S. (2016). PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Research, 44, W16-21.
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., Formsma, K., Gerdes, S., Glass, E. M., & Kubal, M. (2008). The RAST Server: Rapid annotations using subsystems technology. BMC Genomics, 9, 1–15.
Bais, H. P., Fall, R., & Vivanco, J. M. (2004). Biocontrol of Bacillus subtilis against infection of Arabidopsis roots by Pseudomonas syringae is facilitated by biofilm formation and surfactin production. Plant Physiology, 134, 307–319.
Basharat, Z., Yasmin, A., He, T., & Tong, Y. (2018). Genome sequencing and analysis of Alcaligenes faecalis subsp. phenolicus MB207. Scientific Reports, 8, 1–10.
Bayer, P. E., Golicz, A. A., Scheben, A., Batley, J., & Edwards, D. (2020). Plant pan-genomes are the new reference. Nature Plants, 6, 914–920.
Bertani, G. (1951). Studies on lysogenesis I.: The mode of phage liberation by lysogenic Escherichia coli1. Journal of Bacteriology, 62, 293.
Bhardwaj, T., & Somvanshi, P. (2017). Pan-genome analysis of Clostridium botulinum reveals unique targets for drug development. Gene, 623, 48–62.
Blin, K., Shaw, S., Steinke, K., Villebro, R., Ziemert, N., Lee, S. Y., Medema, M. H., & Weber, T. (2019). antiSMASH 5.0: Updates to the secondary metabolite genome mining pipeline. Nucleic Acids Research, 47, W81–W87.
Bóka, B., Manczinger, L., Kocsubé, S., Shine, K., Alharbi, N. S., Khaled, J. M., Münsterkötter, M., Vágvölgyi, C., & Kredics, L. (2019). Genome analysis of a Bacillus subtilis strain reveals genetic mutations determining biocontrol properties. World Journal of Microbiology and Biotechnology, 35, 52.
Bolger, A. M., Lohse, M., & Usadel, B. (2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics, 30, 2114–2120.
Brettin, T., Davis, J. J., Disz, T., Edwards, R. A., Gerdes, S., Olsen, G. J., Olson, R., Overbeek, R., Parrello, B., & Pusch, G. D. (2015). RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Scientific Reports, 5, 8365.
Caulier, S., Nannan, C., Gillis, A., Licciardi, F., Bragard, C., & Mahillon, J. (2019). Overview of the antimicrobial compounds produced by members of the Bacillus subtilis group. Frontiers in Microbiology, 10, 302.
Chaudhari, N. M., Gupta, V. K., & Dutta, C. (2016). BPGA-an ultra-fast pan-genome analysis pipeline. Scientific Reports, 6, 1–10.
Cottingham, R. (2014). The DOE systems biology knowledgebase (KBase). Proceedings of the 5th ACM Conference on Bioinformatics, Computational Biology, and Health Informatics. https://doi.org/10.1145/2649387.2666615
Cui, L., Yang, C., Wei, L., Li, T., & Chen, X. (2020). Isolation and identification of an endophytic bacteria Bacillus velezensis 8–4 exhibiting biocontrol activity against potato scab. Biological Control, 141, 104156.
Cui, W., He, P., Munir, S., He, P., Li, X., Li, Y., Wu, J., Wu, Y., Yang, L., & He, P. (2019). Efficacy of plant growth promoting bacteria Bacillus amyloliquefaciens B9601–Y2 for biocontrol of southern corn leaf blight. Biological Control, 139, 104080.
Dhouib, H., Zouari, I., Ben Abdallah, D., Belbahri, L., Taktak, W., Triki, M. A., & Tounsi, S. (2019). Potential of a novel endophytic Bacillus velezensis in tomato growth promotion and protection against Verticillium wilt disease. Biological Control, 139, 104092.
Dunlap, C., Bowman, M., & Rooney, A. P. (2019). Iturinic lipopeptide diversity in the Bacillus subtilis species group–Important antifungals for plant disease biocontrol applications. Frontiers in Microbiology, 10, 1794.
Fadiji, A. E., & Babalola, O. O. (2020). Elucidating mechanisms of endophytes used in plant protection and other bioactivities with multifunctional prospects. Frontiers in Bioengineering and Biotechnology, 8, 467.
Falcón García, C., Kretschmer, M., Lozano-Andrade, C. N., Schönleitner, M., Dragoŝ, A., Kovács, Á. T., & Lieleg, O. (2020). Metal ions weaken the hydrophobicity and antibiotic resistance of Bacillus subtilis NCIB 3610 biofilms. Npj Biofilms and Microbiomes, 6(1). https://doi.org/10.1038/s41522-019-0111-8
Fashola, M. O., Ngole-Jeme, V. M., & Babalola, O. O. (2016). Heavy metal pollution from gold mines: Environmental effects and bacterial strategies for resistance. International Journal of Environmental Research and Public Health, 13, 1047.
García-Anaya, M. C., Sepulveda, D. R., Sáenz-Mendoza, A. I., Rios-Velasco, C., Zamudio-Flores, P. B., & Acosta-Muñiz, C. H. (2020). Phages as biocontrol agents in dairy products. Trends in Food Science & Technology, 95, 10–20.
Han, N., Qiang, Y., & Zhang, W. (2016). ANItools web: A web tool for fast genome comparison within multiple bacterial strains. Database, 2016.
Hantke, I., Schäfer, H., Janczikowski, A., & Turgay, K. (2019). YocM a small heat shock protein can protect Bacillus subtilis cells during salt stress. Molecular Microbiology, 111, 423–440.
Igiehon, N. O., & Babalola, O. O. (2018). Below-ground-above-ground plant-microbial interactions: Focusing on Soybean, Rhizobacteria and Mycorrhizal Fungi. The Open Microbiology Journal, 12, 261–279.
Jin, P., Wang, H., Tan, Z., Xuan, Z., Dahar, G. Y., Li, Q. X., Miao, W., & Liu, W. (2020). Antifungal mechanism of bacillomycin D from Bacillus velezensis HN-2 against Colletotrichum gloeosporioides Penz. Pesticide Biochemistry and Physiology, 163, 102–107.
Liu, J., Zhou, T., He, D., Li, X.-Z., Wu, H., Liu, W., & Gao, X. (2011). Functions of lipopeptides bacillomycin D and fengycin in antagonism of Bacillus amyloliquefaciens C06 towards Monilinia fructicola. Journal of Molecular Microbiology and Biotechnology, 20, 43–52.
Liu, L., Zhao, X., Huang, Y., Ke, L., Wang, R., & Qi, G. (2020). Protecting tobacco plants from O3 injury by Bacillus velezensis with production of acetoin. Physiologia Plantarum, 170(2), 158–171. https://doi.org/10.1111/ppl.13120
Nurk, S., Bankevich, A., Antipov, D., Gurevich, A. A., Korobeynikov, A., Lapidus, A., Prjibelski, A. D., Pyshkin, A., Sirotkin, A., & Sirotkin, Y. (2013). Assembling single-cell genomes and mini-metagenomes from chimeric MDA products. Journal of Computational Biology, 20, 714–737.
Olanrewaju, O. S. (2016). Isolation of bacterial strains for improved maize production. North-West University (South Africa).
Olanrewaju, O. S., Ayangbenro, A. S., Glick, B. R., & Babalola, O. O. (2018). Plant health: Feedback effect of root exudates-rhizobiome interactions. Applied Microbiology and Biotechnology, 103(3), 1155–1166. https://doi.org/10.1007/s00253-018-9556-6
Olanrewaju, O. S., & Babalola, O. O. (2019). Bacterial consortium for improved maize (Zea mays L.) production. Microorganisms, 7(11), 519. https://doi.org/10.3390/microorganisms7110519
Olanrewaju, O. S., Glick, B. R., & Babalola, O. O. (2017). Mechanisms of action of plant growth promoting bacteria. World Journal of Microbiology and Biotechnology, 33, 197.
Overbeek, R., Olson, R., Pusch, G. D., Olsen, G. J., Davis, J. J., Disz, T., Edwards, R. A., Gerdes, S., Parrello, B., & Shukla, M. (2014). The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST). Nucleic Acids Research, 42, D206–D214.
Peng, G., Zhao, X., Li, Y., Wang, R., Huang, Y., & Qi, G. (2019). Engineering Bacillus velezensis with high production of acetoin primes strong induced systemic resistance in Arabidopsis thaliana. Microbiological Research, 227, 126297.
Richards, P. J., Connerton, P. L., & Connerton, I. F. (2019). Phage biocontrol of Campylobacter jejuni in chickens does not produce collateral effects on the gut microbiota. Frontiers in Microbiology, 10, 476.
Rosier, A., Medeiros, F. H., & Bais, H. P. (2018). Defining plant growth promoting rhizobacteria molecular and biochemical networks in beneficial plant-microbe interactions. Plant and Soil, 428, 35–55.
Rosselli, R., Romoli, O., Vitulo, N., Vezzi, A., Campanaro, S., De Pascale, F., Schiavon, R., Tiarca, M., Poletto, F., & Concheri, G. (2016). Direct 16S rRNA-seq from bacterial communities: A PCR-independent approach to simultaneously assess microbial diversity and functional activity potential of each taxon. Scientific Reports, 6, 1–12.
Tettelin, H., Masignani, V., Cieslewicz, M. J., Donati, C., Medini, D., Ward, N. L., Angiuoli, S. V., Crabtree, J., Jones, A. L., & Durkin, A. S. (2005). Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: Implications for the microbial “pan-genome.” Proceedings of the National Academy of Sciences, 102, 13950–13955.
Torres, M., Llamas, I., Torres, B., Toral, L., Sampedro, I., & Béjar, V. (2020). Growth promotion on horticultural crops and antifungal activity of Bacillus velezensis XT1. Applied Soil Ecology, 150, 103453.
Turan, M., Kitir, N., Alkaya, Ü., Günes, A., Tüfenkçi, S., Yildirim, E., & Nikerel, E. (2016). Making soil more accessible to plants: The case of plant growth promoting Rhizobacteria. Plant Growth. https://doi.org/10.5772/64826
van Heel, A. J., de Jong, A., Song, C., Viel, J. H., Kok, J., & Kuipers, O. P. (2018). BAGEL4: A user-friendly web server to thoroughly mine RiPPs and bacteriocins. Nucleic Acids Research, 46, W278–W281.
Wang, X., Wei, Z., Yang, K., Wang, J., Jousset, A., Xu, Y., Shen, Q., & Friman, V.-P. (2019). Phage combination therapies for bacterial wilt disease in tomato. Nature Biotechnology, 37, 1513–1520.
Woo, O.-G., Kim, H., Kim, J.-S., Keum, H. L., Lee, K.-C., Sul, W. J., & Lee, J.-H. (2020). Bacillus subtilis strain GOT9 confers enhanced tolerance to drought and salt stresses in Arabidopsis thaliana and Brassica campestris. Plant Physiology and Biochemistry, 148, 359–367.
Xie, S. S., Wu, H. J., Zang, H. Y., Wu, L. M., Zhu, Q. Q., & Gao, X. W. (2014). Plant growth promotion by spermidine-producing Bacillus subtilis OKB105. Molecular Plant-Microbe Interactions, 27, 655–663.
Xu, W.-F., Ren, H.-S., Ou, T., Lei, T., Wei, J.-H., Huang, C.-S., Li, T., Strobel, G., Zhou, Z.-Y., & Xie, J. (2019). Genomic and functional characterization of the endophytic Bacillus subtilis 7PJ-16 strain, a potential biocontrol agent of mulberry fruit sclerotiniose. Microbial Ecology, 77, 651–663.
Xu, Z., Zhang, H., Sun, X., Liu, Y., Yan, W., Xun, W., Shen, Q., & Zhang, R. (2019). Bacillus velezensis wall teichoic acids are required for biofilm formation and root colonization. Applied and Environmental Microbiology, 85, e02116-02118.
Yoon, S.-H., Ha, S.-M., Kwon, S., Lim, J., Kim, Y., Seo, H., & Chun, J. (2017). Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. International Journal of Systematic and Evolutionary Microbiology, 67, 1613.
Zeriouh, H., de Vicente, A., Pérez-García, A., & Romero, D. (2014). Surfactin triggers biofilm formation of Bacillus subtilis in melon phylloplane and contributes to the biocontrol activity. Environmental Microbiology, 16, 2196–2211.
Zhang, J., Wang, H., Huang, Q., Zhang, Y., Zhao, L., Liu, F., & Wang, G. (2020). Four superoxide dismutases of Bacillus cereus 0–9 are non-redundant and perform different functions in diverse living conditions. World Journal of Microbiology and Biotechnology, 36, 12.
Acknowledgements
North-West University is gratefully acknowledged for school bursaries to OSO and MSA. ASA appreciates North-West University for a postdoctoral fellowship award. OOB thanks NRF South Africa for funds.
Funding
OOB would like to thank the National Research Foundation, South Africa, for a grant (Ref: UID123634) that has supported research in her laboratory.
Author information
Authors and Affiliations
Contributions
Oluwaseyi Samuel Olanrewaju: conceptualization, methodology, formal analysis, investigation, writing — original draft, writing — review and editing, software, visualization. Modupe Stella Ayilara: methodology, writing — original draft. Ayansina Segun Ayangbenro: methodology, formal analysis, writing — review and editing. Olubukola Oluranti Babalola: conceptualization, investigation, resources, writing — review and editing, supervision, project administration, funding acquisition.
Corresponding author
Ethics declarations
Ethics Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Olanrewaju, O.S., Ayilara, M.S., Ayangbenro, A.S. et al. Genome Mining of Three Plant Growth-Promoting Bacillus Species from Maize Rhizosphere. Appl Biochem Biotechnol 193, 3949–3969 (2021). https://doi.org/10.1007/s12010-021-03660-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-021-03660-3