Abstract
Patterns of gene flow and gene introgression can be used to assess the risk of genetic pollution of wild forest trees from widespread cultivated trees. A comprehensive understanding of the genetic relationships and levels of gene flow among wild and cultivated common walnut (Juglans regia) has become an urgent issue. Using twelve microsatellite markers, we investigated the genetic diversity and gene flow between cultivated and wild trees of J. regia in the Qinling Mountains, China. A high level of genetic variation was detected in both cultivated and wild trees. The mean number of alleles per locus was 17.5. Observed heterozygosity (HO) and expected heterozygosity (HE) were 0.777 and 0.800, respectively. Pollination of mother trees was not by nearest neighbors, and a paternity of 60.7% of offspring evaluated could not be assigned to a local, sampled tree. Pollen flow from cultivated trees to wild trees was infrequent (5.4%), and selfing rates ranged from zero to 25.0%. Male parents were located from 0 to 1005 m from their female partners, with an average pollination distance of 285.1 m. These results are discussed in light of the cultivated species' diversity, outlining the frequent spontaneous genetic contributions from the wild to the cultivated compartment. In addition, the pollen flow parameters provide useful information about the dynamics of pollen movement within J. regia populations.
Similar content being viewed by others
Introduction
Mating patterns, including pollen dispersal, gene flow, and the movement of genes among lineages, play an important role in the evolution of organisms by shuffling the genetic diversity and structure within species (Jones et al. 2008). Genetic invasion between wild species and cultivars by pollen dispersal, seed dispersal, and hybridization has played an important role in plant evolutionary history, especially the domestication of trees as crops (Miller and Gross 2011; Delplancke et al. 2013). Historically, wild species have been used in breeding programs as a source of morphological and genetic variation for plant improvement. In places where crops are grown near their wild ancestors, the reverse movement of genes, from cultivated to wild plants, has important evolutionary consequences for local conspecifics and their relatives (McGranahan et al. 1988; Barbour et al. 2002; Delplancke et al. 2012; Cornille et al. 2013). Because cultivated genotypes can be highly competitive, they can contribute to the local exclusion of wild conspecifics (Trucco et al. 2009). In other cases, cultivated plant species have been selected for non-competitiveness or are less robust because of genetic bottlenecks caused by long-term cultivation (Miller and Gross 2011; Delplancke et al. 2012), but these phenotypes and domestication bottlenecks are often much less pronounced in perennial tree crops.
Hybridization between cultivated and wild trees is facilitated by the frequent absence of an intrinsic reproductive barrier (Gepts and Papa 2003; Cornille et al. 2013). Cultivated to wild gene flow can reduce biodiversity, cause a decline in reproductive fitness, and lead to loss of genetic distinctiveness of native populations (Wolf et al. 2001; Sánchez-de León and Johnson-Maynard 2009). Knowledge of pollen dispersal patterns within forest landscapes is important for managing the integrity of native gene pools by limiting the potential for genetic pollution from non-native plantings.
Walnuts (most Juglans species) belong to a widespread genus of wind-pollinated heterodichogamous species (Bai et al. 2006, 2007; Kimura et al. 2012; Zhao et al. 2018). The common walnut (also called the English or Persian walnut, J. regia L.) is a monoecious wind-pollinated deciduous tree native to Eurasia but now grown in many countries with a temperate climate (Manning 1978; Gunn et al. 2010; Aradhya et al. 2017). It has been cultivated for its timber and nuts for at least 6800 years (Beer et al. 2008; Feng et al. 2018). In many parts of Asia it is difficult to know if autochthonous trees are wild or feral (Pollegioni et al. 2015; Aradhya et al. 2017; Bernard et al. 2018) as there are no obvious phenotypic indicators of domestication. Nuts from apparently wild trees are harvested by villagers who transport them to markets where they may be sold alongside locally grown improved varieties. Because common walnut is long-lived and anemophilous, gene flow between wild trees and modern cultivated genotypes or their seedlings is likely, even in areas that may be centers of diversity.
We investigated gene flow between (apparently) wild and cultivated common walnut trees in the Qinling Mountains of China, a region believed to contain an ancient population of J. regia (Cheng et al. 2013; Guo et al. 2013). Our goals were to: (1) Characterize the genetic diversity of common walnuts (J. regia) in the Qinling Mountains; (2) Investigate the relative genetic contribution of cultivated J. regia to the seeds of wild trees in the Qinling Mountains; and, (3) Determine the pollen dispersal distance at research locations.
Materials and methods
Sampling and plant materials
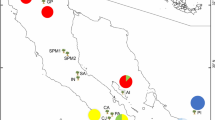
Common walnut is valued for both nuts and timber. To characterize the genetic diversity of cultivated common walnuts, we sampled seeds from trees planted by farmers in 16 locations around the Qinling Mountains, Shaanxi province, China (Table 1, Fig. 1). For clarity, these cultivated trees are described as “cultivars” even though they were not grafted but instead chance seedlings or seedlings of locally desirable genotypes. At sites with cultivated trees, a sample of genotypes were collected that were traditionally cultivated by local farmers who had no access to modern, commercial cultivars. To investigate the relative genetic contribution of cultivated J. regia to the seeds of wild trees, we sampled 275 seeds from 19 mother trees at a single location (Table 1; Fig. 1). Both the seeds and the mother trees were genotyped at 12 loci and a parentage analysis used to determine the percent of offspring derived from self-pollinations, pollinations from nearby mother trees, pollinations from trees outside the study area that were not genotyped, and pollinations from the closest cultivated population (HH and II; Fig. 1). To determine the pollen dispersal distance (Table 1, Fig. 1), the sample locations were mapped using GIS software ArcGIS (version 10.0; ESRI 2010).
Locations of genotyped coomon walnut tree within the Qinling Mountains. The numbers indicate the number of samples genotyped. Numbers 1–16 indicate 16 locations of cultivar trees, while the 19 wild mother trees are from A to S in the box of red dotted line (The numbers of seeds for each tree used for data analysis, details see Table 1). The number of the different scales on the map. QH = Qinghai Province, GS = Gansu Province, SX = Shaanxi Province, SXI = Shanxi Province, HN = Henan Province, SC = Sichuan Province, HB = Hubei Province, CS = Chongqing Direct-controlled Municipality
SSR genotyping
DNA was extracted from leaf and seed samples using the methods of Doyle and Doyle (1987) and Zhao and Woeste (2011). Twelve microsatellite loci (Woeste et al. 2002; Victory et al. 2006; Ahmed et al. 2012; Pollegioni et al. 2015) (Table 2) were used for genotyping. Each forward primer was marked with fluorescent dye (FAM, TAMRM, HEX and ROX). The PCR products were sent to Shagon Biotech Life Science Products and Services for analysis. The genetic analysis process was similar to the methods in Feng et al. 2018.
Microsatellite data analysis
Genemapper software (Applied Biosystem, Foster City, CA, USA) was used to score all genotype data. After removing 18 samples of seeds of cultivated individuals (seeds) and 36 samples of seeds of wild individuals (seeds) that had genotypes that did not match their presumed maternal parent based on CERVUS v3.0 (Kalinowski et al. 2007), a total of 317 individuals were retained for analysis. The exclusion probabilities for one parent and two parents was 80%. We tested for evidence of null alleles using MICRO-CHECKER (Van Oosterhout et al. 2004).
Deviations per locus and per mother tree from Hardy-Weinberg equilibrium (HWE) were tested using GENEPOP (Raymond and Rousset 1995) and Arlequin 3.5 (Excoffier and Lischer 2010). CERVUS v3.0 software (Kalinowski et al. 2007) and POPGEN version 1.32 (Yeh and Boyle 1996) were used to calculate the genetic diversity parameters, including percent polymorphic loci (95% criterion), the total number of alleles (Na), effective number of alleles (Ne), allelic richness (Ar), gene diversity (HS), and overall gene diversity (HT). The observed (HO) and expected (HE) heterozygosities were calculated based on the allele frequencies using POPGENE (Yeh and Boyle 1996).
Genetic differentiation between groups (FST) and linkage disequilibrium (LD) between loci were tested using the program GENEPOP v4.0 (Raymond and Rousset 1995). The significance of difference between genetic groups, measured as FST, was determined by permutation tests (10,000) using Arlequin v3.5 (Excoffier and Lischer 2010). RST was calculated using SPAGEDI (Hardy and Vekemans 2002). We performed a two-level (among trees and with trees) analysis of molecular variance (AMOVA) based on 239 seed from 19 wild mother trees using ARLEQUIN v3.5 (Excoffier and Lischer 2010).
The overall genetic variation among cultivated and wild trees was represented using principal coordinate analysis (PCoA) using GenALEx v6.5 (Peakall and Smouse 2012). To evaluate the admixture and genetic populations within wild trees only, a genetic structure analysis of 239 wild seeds was performed using a model-based Bayesian clustering of individuals, as implemented in STRUCTURE v2.3.4 (Pritchard et al. 2000). We also add the cultivated seeds to the STRUCTURE analysis to check the genetic mixture. This software uses a Markov Chain Monte Carlo (MCMC) framework in which the algorithm explores a parameter space considering individual admixture proportion, locus-specific ancestries, population allele frequencies, and the expected admixture of the data set, assuming a user-defined K number of groups. STRUCTURE was run using 100,000 burn-in MCMC iterations, with run lengths of 1,000,000 iterations, and ten replicates per run for K = 1–15 clusters, with admixture (Pritchard et al. 2000). STRUCTURE HARVESTER was used to calculate the optimal value of K (Earl and Vonholdt. 2012) using the deltaK criterion (Evanno et al. 2005), and the inferred clusters were drawn as colored box-plots using program DISTRUCT (Rosenberg 2004).
Maternity and paternity analysis
A total of 275 seeds from 19 adult wild walnut trees were analysed for maternity using CERVUS v3.0 (Kalinowski et al. 2007), a software based on maximum-likelihood methods. The simulation parameters were: 10,000 cycles, 19 candidate mother trees, 19/275 as the proportion of candidate mothers sampled, 0.975 as the proportion of loci typed, 0.01 as the proportion of mistyped, 0.01 error rate in likelihood calculations, 0.80 for the relaxed confidence level and 0.95 for the strict confidence level. After verification of maternal parentage for each offspring, the number of candidate male parents specified in the simulation was the approximate size of the male gene pool for the cultivated and wild trees in this study, which included both sampled and unsampled male parents. The total of number of identified male parents was 371 and the proportion of male parents sampled were adjusted to 0.80, 0.05 as the proportion of mistyped, 0.05 error rate in likelihood calculations, 0.65 for the relaxed confidence level and 0.95 for the strict confidence level.
Mating pattern and pollen dispersal
The relatedness among all 19 adult wild trees was calculated using software RELATEDNESS v5.0.5 (Goodnight and Queller 1999). Observed distances of pollen migration were used to construct a dispersal curve to describe pollen movement within the population. Measurements consisted of the distance between the mother and the putative father when the most likely father was assigned. In addition, the distances between each mother tree and all other candidate fathers were also measured to determine whether the observed inter-mate distance was influenced by the spatial arrangement of the adult trees. Gene flow (Nm) was estimated using Arlequin v3.11 (Excoffier and Lischer 2010) and MIGRATE v3.6 (Beerli 2006).
Results
Genetic variation and structure of wild walnut seeds and cultivars as revealed by microsatellite data
Estimates of genetic diversity varied among microsatellite loci and among the wild offspring cohorts and adult (wild) trees [N = 18 after removal of wild parent Tree-N (Fig. 1) due to poor DNA quality]. Among wild samples (N = 239), alleles per locus (Na) ranged from 9 to 34, with a total of 192 alleles across the 12 loci. The mean number of alleles per locus was 16.0. The mean polymorphic information content (PIC) was 0.774 (Table 2). Allelic richness (Ar) estimated among all trees ranged from 4.00 to 9.06 with an average of 6.85. Observed heterozygosity (HO) among loci was 0.803, with a range of 0.548–0.927. Expected heterozygosity (HE) among loci was 0.796, with a range of 0.563–0.957 (Table 2).
Among samples from cultivated trees (78 seeds from 16 locations), the range of alleles per locus (Na) was from 7 to 29, with a total of 187 alleles across the 12 loci. The mean number of alleles per locus was 15.58. The mean polymorphic information content (PIC) was 0.779 (Table 3). Observed heterozygosity (HO) among loci was 0.672 with a range of 0.377–0.880. The mean expected heterozygosity (HE) among loci was 0.838, with a range of 0.634–0.955. The mean polymorphic information content (PIC) was 0.794 and the mean effective number of alleles (Ne) was 1.93 with a range of 1.00–4.00 (Table 3). In wild trees, by contrast, the mean effective number of alleles (Ne) was 3.41 with a range of 2.15–4.85 (Table 2). Microsatellites revealed clear genetic differentiation between the 239 wild progenies and 78 cultivated progenies (Figs. 2 and 3). Both wild populations and cultivars were distinguished by the first two coordinates of the principal coordinate analyses (Fig. 2, accounting for 63.4% of the observed variance).
Genetic structure
Bayesian clustering of 239 progenies from 18 wild mother trees identified three populations as most likely (Fig. 4). These populations are distinguishable despite relatively high gene flow among the wild mother trees (Nm = 1.534). Visual examination of the spatial arrangement of the populations within the site from which the wild seeds were collected reveals no obvious spatial association among the trees of each population (Fig. 1). The wild populations were genetically distinct from the seeds from cultivated trees (Fig. 3), which all belonged to a common population.
Maternity and paternity analysis
Paternity analysis of 371 seeds revealed that the male parent of 94 (25.3%) samples could be identified based on the genotypes of the seeds and their presumed female parent. Of these, 36 samples (15.1%) were seeds from cultivated trees, 61 (25.5%) were from wild adult mother trees with 80% confidence (Table 4). Among wild trees, mean distance between successful males and their female mates was 285.1 m, ranging from 61.2 to 1005.5 m, not including self-pollinations. Most successful pollinizers (75%) were from 200 to 600 m from their mate, and only three trees were more than 1250 m from their mate (Fig. 5). The rate of gene flow (0.425) between wild and cultivated J. regia was estimated using FST, which was 0.088, ranged from 0.046 (GG-LL) and from 0.147 (FF-NN), which was lower than the average FST among 18 progenies of wild trees (FST = 0.094), ranged from 0.053 (J-K) and from 0.137 (C-I). The genetic differentiation (FST) was 0.042 between wild and cultivated groups; AMOVA revealed that most of the genetic variation (96%) was within wild or cultivated trees (Table 5). Paternity analysis showed that none of the wild mother trees was father of a seed from a cultivated tree, although coalescent-based estimations of gene flow (Nm = 1.48 among wild mother trees; Nm = 2.58 among cultivated trees, Nm = 1.76 between wild and cultivated trees) indicated that cultivated J. regia was expected to contributed about 13 migrants (5.4%) to the seeds of the wild progenies (Fig. 6).
Distribution of J. regia trees in the study population showing the locations of eighteen wild mother trees. Small circles and pie graphs indicate the profiles of genotyped progeny arrays from each mother tree for seed lot collection from 2011. Large letters around the pie indicate the wild mother tree’s name. Dark gray, white, light gray, and black wedges represent selfing, male parent was a cultivated plant, two parents within study site, and unassigned seeds, respectively (details see Table 5)
Mating system and pollen dispersal
Of 239 progeny samples, 36 (15.1%) were determined to have arisen from self-pollination. Among wild mother trees, the selfing rate varied between zero to 16.7% (Table 6; Fig. 6). The percentage of seeds with two wild parents within the study site varied among mother trees from zero to 38.5% (Table 6). In addition, 145 progenies (60.7%) were not assigned pollen parents (paternity analysis did not match any sampled wild or cultivated genotype). These were most likely the result of immigrant pollen from outside the study area or were sired by un-sampled trees within the study site. Contamination rates for individual wild mother trees ranged from 50.0% to 91.7% (Table 6).
Discussion
Performance of microsatellite markers in genetic variation and genetic structure of walnut
Microsatellite markers provided robust and highly informative signals based on high levels of polymorphism (Woeste et al. 2002; Dangl et al. 2005; Victory et al. 2006; Pollegioni et al. 2015). In general, it is expected that the genetic diversity of wild trees would be higher than cultivated trees. Our investigation revealed genetic variation in progenies of wild and cultivated J. regia in the Qinling Mountains were similar, although there were some important differences, and wild and cultivated trees belonged to distinct genetic groups (Fig. 3). Cultivated and wild trees were similar in terms of number of alleles (Na), expected heterozygosity (HE), and polymorphic information content (PIC) (Tables 2 and 3). In contrast, cultivated trees and wild trees differed in terms of effective number of alleles (Ne) (1.93 versus 3.41) and observed heterozygosity (HO) (0.672 versus 0.803) (Tables 2 and 3). These results indicate that the cultivated trees and wild trees differed considerably in two important measures of diversity, possibly because of human selection, although it is likely that most cultivated trees in the Qinling Mountains are only one or two generations removed from a wild population.
Contributions of wild walnuts to cultivated trees
These results are in accordance with studies of other plants that revealed substantial genetic exchange between wild and cultivated varieties of the same species. For example, patterns with a similar trend were observed for maize (Matsuoka et al. 2002), apples (Coart et al. 2006; Cornille et al. 2012), cultivated olive trees (Breton et al. 2008), and grape cultivars (Myles et al. 2011).
Wild walnut trees have long been used as a source of genetic novelty in breeding programs (Pollegioni et al. 2015). On the other hand, these results raised a question of whether wild-to-cultivated exchanges remained exclusively spontaneous or whether traditional practices could have facilitated the fixation of introgressed wild genes into cultivated walnut trees. For Juglans, several authors reported the direct use of wild species as rootstocks (McGranahan and Leslie 2009). Additionally, germplasm resources are rich and the varieties numerous in J. regia which may be caused by a long cultivation history, and many are biologically and economically important because of their high-quality timer and nutritious nuts. Traditional practices in walnuts breeding remain largely unknown s (Rogers 2004; Beer et al. 2008).
Nuclear microsatellites (SSRs) showed that genetic exchanges between cultivated J. regia and wild J. regia were directional (from cultivar to wild), and outlined substantial cultivated-to-wild gene flow (Nm = 0.4252), confirming that J. regia genes could spontaneously be introgressed into their wild resources. And this genetic transfer can have significant evolutionary consequences, especially if the inserted transgene is adaptive under natural conditions (Felber et al. 2007; Sun et al. 2019). For example, traits such as enhanced fertility (Zhao and Woeste 2011), late flowering (Cornille et al. 2012), and antifungal (e.g., S. clavigignenti-juglandacearum, Ross-Davis et al. 2008) could favor the emergence of highly competitive phenotypes if transferred into wild species. Our results also highlighted the importance when inferring the origins of walnut domestication using molecular genetic data, because gene flow from wild walnuts can have a bias on distance-based ancestry inferences (Cornille et al. 2012; De Andrés et al. 2012). Finally, this study revealed that cultivated-to-wild gene flow occurred commonly among the walnuts located in the Qinling Mountains. There are no barriers preventing gene flow between wild and cultivated walnut trees at the current time in the study site. Further, these results suggest that transgenes could potentially introgress from cultivated into wild Persian walnut populations, demonstrating the need for detailed characterization of cultivated to wild gene flow within the Juglans species complex (Yuan et al. 2018; Dang et al. 2019).
Mating system, out crossing rates, and pollen dispersal
The mating system of walnuts is characterized by a protogynous–protandrous dimorphism which is a form of heterodichogamy (Bai et al. 2007; Kimura et al. 2012) intended to avoid self-pollination. A genetic model of this system predicts a stable polymorphism with both mating types in equal proportion in a randomly mating population (Härdling and Bergsten 2006). The proportion of progeny derived from self-pollinations in our study was 15.1%, less than three times as outside trees. This selfing rate was considerably higher than rates reported in other Juglans species (Bai et al. 2007) and in other wind-pollinated species (Kimura et al. 2012). The rate of self-pollination can vary over years and among populations and may be strongly affected by local factors (Kery et al. 2003). Bai et al. (2007) showed that self- mating was rare in a high-density, morph-ratio-unbiased population of the heterodichogamous Manchurian walnut, Juglans mandshurica Maxim. Kimura et al. (2012) reported that the selfing rate was 13.8% of Japanese walnut (J. ailantifolia Carr.) in a 2.5 ha study site. The population was wild, low density, and morph-ratio-biased. In our study, selfing rate varied between zero to 25.0% with a mean of 15.1% (Fig. 6); the highest rates of selfing were observed in tree-M and tree-H; Fig. 6). Moreover, the un-assigned wild trees impact the selfing rate caused by the missing data in this study. The simplest explanation for variance in selfing rates among trees is difference in the availability of mating partners in the neighbourhood, but factors related to phenology likely play a role, especially within-plant bloom overlap. Therefore, a relatively lengthy intra-morph bloom overlap may have affected the extent of assortative mating in this study. The inbreeding coefficient for the seeds of the wild parents was 0.077.
Averaged across all seeds, the mean distance between parents (d) was 285.1 m for wild-mother trees. Immigration rates were approximately 60.9%. These findings indicate that pollination by distant males may be relatively common in this population (Fig. 5). Kimura et al. (2012) showed that distance to pollinizers in a wild population of Juglans ailantifolia was 392 m and 233 m for protogynous and protandrous mother trees, respectively. Bai et al. (2007) showed that pollen dispersal in J. mandshurica was predominantly short-distance, and local density of reproductive trees affected the patterns of pollen dispersal in forest trees. Mating patterns were thus also affected by both density and tree size (Jolivet et al. 2012).
Our study revealed that in this population of J. regia in the Qinling Mountains, the longest pollination distance was 1.0 km. Large-scale studies are needed to disentangle relative influences of these factors on the mating system and pollination success (Jolivet et al. 2012; Davies et al. 2015).
Conclusions
Pollination of mother trees was outcrossed, often by neighbors, and pollinizers outside the immediate sampled area accounted for about 60.7% of the sampled seeds. In a few cases, pollination of wild trees was by nearby cultivated trees (5.4%). The selfing rate among the wild trees varied from zero to 25.0%. Pollinizers ranged from zero (self-pollinations) to 1005 m from their respective female partners, with an average distance of 285.1 m. This description of pollen flow provides useful information about the dynamics of pollen movement within wild J. regia populations.
References
Ahmed N, Mir JI, Mir RR, Rather NA, RashidR WSH, Mir H, Sheikh MA (2012) SSR and RAPD analysis of genetic diversity in walnut (Juglans regia L.) genotypes from Jammu and Kashmir, India. Physiol Mol Biol Plants 18(2):149–160
Aradhya M, Velasco D, Ibrahimov Z, Toktoraliev B, Maghradze D, Musayev M, Bobokashvili Z, Preece JE (2017) Genetic and ecological insights into glacial refugia of walnut (Juglans regia L.). PLoS One 12(10):e0185974
Bai WN, Zeng YF, Liao WJ, Zhang DY (2006) Flowering phenology and wind-pollination efficacy of heterodichogamous Juglans mandshurica (Juglandaceae). Ann Bot 98:397–402
Bai WN, Zeng YF, Zhang DY (2007) Mating patterns and pollen dispersal in a heterodichogamous tree, Juglans mandshurica (Juglandaceae). New Phytol 176(3):699–707
Barbour RC, Potts BM, Vaillancourt RE, Tibbits WN, Wiltshire RJ (2002) Gene flow between introduced and native Eucalyptus species. New For 23(3):177–191
Beer R, Kaiser F, Schmidt K, Ammann B, Carraro G, Grisa E, Tinner W (2008) Vegetation history of the walnut forests in Kyrgyzstan (Central Asia): natural or anthropogenic origin? Quat Sci Rev 27(5–6):621–632
Beerli P (2006) Comparison of Bayesian and maximum-likelihood inference of population genetic parameters. Bioinformatics 22(3):341–345
Bernard A, Barreneche T, Lheureux F, Dirlewanger E (2018) Analysis of genetic diversity and structure in a worldwide walnut (Juglans regia L.) germplasm using SSR markers. PLoS One 13(11):e0208021
Breton C, Pinatel C, Médail F, Bonhomme F, Bervillé A (2008) Comparison between classical and Bayesian methods to investigate the history of olive cultivars using SSR-polymorphisms. Plant Sci 175(4):524–532
Cheng F, Peng X, Zhao P, Yuan J, Zhong C, Cheng Y, Cui C, Zhang S (2013) Soil microbial biomass, basal respiration and enzyme activity of main forest types in the Qinling Mountains. PLoS One 8(6):e67353
Coart E, Van Glabeke S, De Loose M, Larsen AS, RoldánRuiz I (2006) Chloroplast diversity in the genus Malus: new insights into the relationship between the European wild apple (Malus sylvestris (L.) Mill.) and the domesticated apple (Malus domestica Borkh.). Mol Ecol 15(8):2171–2182
Cornille A, Gladieux P, Smulder MJ, Roldán-Ruiz I, Laurens F, Le Cam B, Nersesyan A, Clavel J, Olonova M, Feugey L, Gabrielyan I, Zhang X, Tenaillon ML, Giraud T (2012) New insight into the history of domesticated apple: secondary contribution of the European wild apple to the genome of cultivated varieties. PLoS Genet 8(5):e1002703
Cornille A, Giraud T, Bellard C, Tellier A, Cam BL, Smulders MJM, Kleinschmit J, Roldan-Ruiz I, Gladieux P (2013) Post-glacial recolonization history of the European crabapple (Malus sylvestris Mill.), a wild contributor to the cultivated apple. Mol Ecol 22(8):2249–2263
Dang M, Yue M, Zhang M, Zhao G, Zhao P (2019) Gene introgression among closely related species in sympatric populations: a case study of three walnut (Juglans) species. Forests 10(11):965
Dangl GS, Woeste K, Aradhya MK, Koehmstedt A, Simon C, Potter D, Leslie CA, McGranahan G (2005) Characterization of 14 microsatellite markers for genetic analysis and cultivar identification of walnut. J Am Soc Hortic Sci 130(3):348e354
Davies SJ, Cavers S, Finegan B, White A, Breed MF, Lowe AJ (2015) Pollen flow in fragmented landscapes maintains genetic diversity following stand-replacing disturbance in a neotropical pioneer tree, Vochysia ferruginea Mart. J Hered 115(2):125–129
De Andrés MT, Benito A, Pérez-Rivera G, Ocete R, Lopez MA, Gaforio L, MuozUÑOZ G, Cabello F, Martínez-Zapter JM, Arroyo-García R (2012) Genetic diversity of wild grapevine populations in Spain and their genetic relationships with cultivated grapevines. Mol Ecol 21(4):800–816
Delplancke M, Alvarez N, Espíndola A, Joly H, Benoit L, Brouck E, Arrigo N (2012) Gene flow among wild and cultivated almond species: insights from chloroplast and nuclear markers. Evol Appl 5(4):317–329
Delplancke M, Alvarez N, Benoit L, Espíndola AI, Joly H, Neuenschwander S, Arrigo N (2013) Evolutionary history of almond tree domestication in the Mediterranean basin. Mol Ecol 22(4):1092–1104
Doyle JJ, Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11–15
Earl DA, Vonholdt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4(2):359–361
ESRI (2010) ArcGIS version 10.0. Environmental Systems Research Institute, Redlands
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14(8):2611–2620
Excoffier L, Lischer HEL (2010) Arlequin suite version 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10(3):564–567
Felber F, Kozlowski G, Arrigo N, Guadagnuolo R (2007) Genetic and ecological consequences of transgene flow to the wild flora. Adv Biochem Eng Biotechnol 107(1):173–205
Feng X, Zhou H, Zulfiqar S, Luo X, Hu Y, Feng L, Malvolti ME, Woeste K, Zhao P (2018) The phytogeographic history of common walnut in China. Front Plant Sci 9:1399
Gepts P, Papa P (2003) Possible effects to (trans) gene flow from crops on the genetic diversity from landraces and wild relatives. Environ Biosaf Res 2(2):89–103
Goodnight KF, Queller DC (1999) Computer software for performing likelihood tests of pedigree relationship using genetic markers. Mol Ecol 8(7):1231–1234
Gunn BF, Aradhya M, Salick JM, Miller AJ, Yang Y, Lin L, Xian H (2010) Genetic variation in walnuts (Juglans regia and J. sigillata; Juglandaceae): species distinctions, human impacts, and the conservation of agrobiodiversity in Yunan, China. Am J Bot 97(4):660–671
Guo Y, Li G, Hu Y, Kang D, Wang D, Yang G (2013) Regeneration of Betula albosinensis in strip clearcut and uncut forests of the Qinling Mountains in China. PLoS One 8(3):e59375
Härdling R, Bergsten J (2006) Nonrandom mating preserves intrasexual polymorphism and stops population differentiation in sexual conflict. Am Nat 167:401–409
Hardy OJ, Vekemans X (2002) SPAGeDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes 2(4):618–620
Jolivet C, Höltken AM, Liesebach H, Steiner W, Degen B (2012) Mating patterns and pollen dispersal in four contrasting wild cherry populations (Prunus avium L.). Eur J For Res 131(4):1055–1069
Jones ME, Shepherd M, Henry R, Delves A (2008) Pollen flow in Eucalyptus grandis determined by paternity analysis using microsatellite markers. Tree Genet Genomes 4(1):37–47
Kalinowski ST, Taper ML, Marshall TC (2007) Revising How the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Mol Ecol 16(5):1099–1006
Kery M, Matthies D, Schmid B (2003) Demographic stochasticity in population fragments of the declining distylous perennial Primula veris (Primulaceae). Basic Appl Ecol 4(3):197–206
Kimura MK, Goto S, Suyama Y, Matsui M, Woeste K, Seiwa K (2012) Morph-specific mating patterns in a low-density population of a heterodichogamous tree, Juglans ailantifolia. Plant Ecol 213(9):1477–1487
Manning WE (1978) The classification within the Juglandaceae. Ann Mo Bot Gard 65(4):1058–1087
Matsuoka Y, Yamazaki Y, Ogihara Y, Tsunewaki K (2002) Whole chloroplast genome comparison of rice, maize, and wheat: implications for chloroplast gene diversification and phylogeny of cereals. Mol Evol 19(12):2084–2091
McGranahan G, Leslie C (2009) Breeding Walnuts (Juglans Regia). In: Jain SM, Priyadarshan PM (eds) Breeding plantation tree crops: temperate species. Springer Sciencet Business Media, LLC, pp 268–270
McGranahan GH, Hansen J, Shaw DV (1988) Inter- and intraspecific variation in California black walnuts. J Am Soc Hortic Sci 83(5):210–215
Miller AJ, Gross BL (2011) From forest to field: perennial fruit crop domestication. Am J Bot 98(9):1389–1414
Myles S, Boyko AR, Owens CL, Brown PJ, Grassi F, Aradhya MK, Prins B, Reynolds A, Chia J, Ware D, Bustamante CD, Buckler ES (2011) Genetic structure and domestication history of the grape. Proc Natl Acad Sci USA 108(9):3530–3535
Peakall R, Smouse PE (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research—an update. Bioinformatics 28(19):2537–2539
Pollegioni P, Woeste KE, Chiocchini F, Del Lungo S, Olimpieri I, Tortolano V, Clark J, Hemery GE, Mapelli S, Malvolti ME (2015) Ancient humans influenced the current spatial genetic structure of common walnut populations in Asia. PLoS One 10(9):e0135980
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155(2):945–959
Raymond M, Rousset F (1995) GENEPOP (Version 1.22): population genetics software for exact tests and ecumenicism. J Hered 86(3):248–249
Rogers R (2004) Temperate ecosystems, Juglandaceae. Elsevier Ltd, Amsterdam, Holland, pp 1427–1430
Rosenberg NA (2004) DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes 4(1):137–138
Ross-Davis A, Huang Z, Mckenna J, Ostry M, Woeste K (2008) Morphological and molecular methods to identify butternut (Juglans cinerea) and butternut hybrids: relevance to butternut conservation. Tree Physiol 28(7):1127–1133
Sánchez-de León Y, Johnson-Maynard J (2009) Dominance of an invasive earthworm in native and non-native grassland ecosystems. Biol Invasions 11(6):1393–1401
Sun Y, Hou N, Woeste K, Zhang C, Yue M, Yuan X, Zhao P (2019) Population genetic structure and adaptive differentiation of iron walnut Juglans regia subsp. sigillata in southwestern China. Ecol Evol 9:14154–14166
Trucco F, Tatum T, Rayburn AL, Tranel PJ (2009) Out of the swamp: unidirectional hybridization with weedy species may explain the prevalence of Amaranthus tuberculatus as a weed. New Phytol 184(4):819–827
Van Oosterhout C, Hutchinson WF, Wills DP, Shipley P (2004) MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Resour 4(3):535–538
Victory ER, Glaubitz JC, Rhodes OE, Woeste KE (2006) Genetic Homogeneity in Juglans nigra (Juglandaceae) at nuclear microsatellites. Am J Bot 93(1):118–126
Woeste K, Burns R, Rhodes O, Michler C (2002) Thirty polymorphic nuclear microsatellite loci from black walnut. J Hered 93(1):58–60
Wolf DE, Takebayashi N, Riesberg LH (2001) Predicting the risk of extinction through hybridization. Conserv Biol 15(4):1039–1053
Yeh FC, Boyle TB (1996) Population genetic analysis of codominant and dominant markers and quantitative traits. Belg J Bot 129:157
Yuan X, Sun Y, Bai X, Dang M, Feng X, Zulfiqar S, Zhao P (2018) Population structure, genetic diversity, and gene introgression of two closely related walnuts (Juglans regia and J. sigillata) in southwestern China revealed by EST-SSR markers. Forests 9(10):646
Zhao P, Woeste K (2011) DNA markers identify hybrids between butternut (Juglans cinerea L.), and Japanese walnut (Juglans ailantifolia Carr.). Tree Genet Genomes 7(3):511–533
Zhao P, Zhou HJ, Potter D, Hu YH, Feng XJ, Dang M, Feng L, Zulfiqar S, Liu WZ, Zhao GF, Woeste K (2018) Population genetics, phylogenomics and hybrid speciation of Juglans in China determined from whole chloroplast genomes, transcriptomes, and genotyping-by-sequencing (GBS). Mol Phylogenet Evol 126:250–265
Acknowledgements
The authors wish to thank Hailong Xia, Qi Xu, Jia Yang, and Tao Zhou for sample collection. Mention of a trademark, proprietary product, or vendor does not constitute a guarantee or warranty of the product by the U.S. Department of Agriculture and does not imply its approval to the exclusion of other products or vendors that also may be suitable.
Author information
Authors and Affiliations
Corresponding author
Additional information
Corresponding editor: Yanbo Hu.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Project funding: This work was supported by Qinling National Forest Ecosystem Research Station, National Natural Science Foundation of China (Nos. 31200500, 41471038, and 32070372), Shaanxi Academy of Science Research Funding Project (Y19Z604F12), Natural Science Foundation of Shaanxi Province of China (2019JM-008), and the public health specialty in the department of traditional Chinese Medicine (2017-66, 2018-43, 2019-68).
The online version is available at http://www.springerlink.com.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Zhou, H., Zhao, P., Woeste, K. et al. Gene flow among wild and cultivated common walnut (Juglans regia) trees in the Qinling Mountains revealed by microsatellite markers. J. For. Res. 32, 2189–2201 (2021). https://doi.org/10.1007/s11676-020-01254-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11676-020-01254-z