Abstract
Single-cell analysis is a powerful technology that has found widespread use in recent years. For diseases with involvement of adaptive immunity, single-cell analysis of antigen-specific T cells and B cells is particularly informative. In autoimmune diseases, the adaptive immune system is obviously at play, yet the ability to identify the culprit T and B cells recognizing disease-relevant antigen can be difficult. Celiac disease, a widespread disorder with autoimmune components, is unique in that disease-relevant antigens for both T cells and B cells are well defined. Furthermore, the celiac disease gut lesion is readily accessible allowing for sampling of tissue-resident cells. Thus, disease-relevant T cells and B cells from the gut and blood can be studied at the level of single cells. Here we review single-cell studies providing information on such adaptive immune cells and outline some future perspectives in the area of single-cell analysis in autoimmune diseases.
Similar content being viewed by others
Introduction
Single-cell analysis has revolutionized many research fields, including studies of adaptive immune responses and autoimmunity. Here we will review such advances in the context of a particular disorder—celiac disease (CeD). We have chosen to structure this review around some common single-cell approaches used to dissect adaptive immune responses, and we start by giving a brief general overview of such approaches. We subsequently describe CeD with its clinical and immunological features. This common disorder, caused by a harmful immune response to cereal gluten proteins, presents with many autoimmune characteristics. We provide a systematic review of how each single-cell approach has advanced the understanding of the role of adaptive immune cells in this disease, before giving some future perspectives on single-cell analysis for other autoimmune diseases, highlighting lessons we have learned from studies of CeD.
Single-cell approaches to dissect adaptive immune responses: general principles
Detection of antigen-specific cells
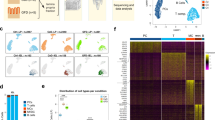
Knowledge about the antigens being targeted in an immune response allows for the isolation of antigen-specific T cells and B cells using T- and B-cell receptor (TCR and BCR) binding ligands as baits (Fig. 1a). B cells can be detected with monovalent or multivalent BCR ligands.1 However, due to low TCR-ligand affinities, the detection of T cells only became feasible with multimer technologies that increase the receptor avidity.2,3
a Antigen-specific T and B cells can be labeled using antigen-ligands as bait and visualized in situ by immunohistochemistry or ex vivo by flow cytometry or mass cytometry. b Various protocols are available for single-cell sequencing of antigen immune receptor repertoires from genomic DNA or RNA, yielding paired, full-length T-cell receptor (TCR) or B-cell receptor (BCR) sequences. c The BCR of single B cells can be expression cloned, and the resulting human monoclonal antibodies can be used to study their specificity, affinity, and binding mode. d BCR sequences may be used as input to computational tools for creation of phylogenetic trees to track evolving clones. e Flow cytometry analysis of antibody-labeled cells enables phenotyping of millions of single cells as well as sorting of single cells into plates for downstream analyses. f Labeling of cells with metal-conjugated antibodies allows for phenotyping of around 40 markers by mass cytometry. g Single-cell RNA-sequencing facilitates unbiased clustering of cells into populations based on the full transcriptional state of each cell.
Single-cell antigen immune receptor sequencing
The fast-developing field of sequencing technologies has revolutionized profiling of TCR- and BCR-repertoires through deep sequencing of amplified RNA or DNA from the variable (V)-region of immune receptor loci (AIRR-seq). Single-cell AIRR-seq (scAIRR-seq) adds chain-pairing information and enables enumeration of cells with identical receptors (Fig. 1b). Expression cloning of human monoclonal antibodies (hmAbs) from single disease-relevant plasma cells (PCs)4 facilitates studies of binding strength, specificity, target epitopes and binding mode (Fig. 1c). Furthermore, full-length, paired TCR- or BCR-sequences makes it possible to study interaction of epitopes and paratopes, for instance by mutational and structural studies.
Lineage tracing
Since TCRs and BCRs are highly diverse molecules created by somatic rearrangements, they can be used to track ancestors of a single T cell or B cell, termed clones (Fig. 1d). Thus, identification of identical TCRs at different time points and tissues indicates a systemic, long-lasting immune response toward the target antigen. As B-lineage cells undergo somatic hypermutation (SHM), clonally related B-lineage cells have similar, but not necessarily identical, BCRs. Acquisition of mutations during affinity maturation can be computationally reconstructed into phylogenetic trees and analyzed at the clone or repertoire level.5,6 Paired chain data from single cells gives the most accurate representation of lineage trees.
Flow cytometry
Flow cytometry is an invaluable tool to study cell-surface molecules and intracellular targets in a large number of single cells (Fig. 1e). Currently, more than 20 markers may be analyzed simultaneously by conventional flow cytometry.7 Flow cytometry allows for rapid analysis of many million cells. Importantly, analyzed cells can be sorted and used for downstream analyses. However, some prior knowledge on marker expression is needed, and measurement of several markers at once requires correct compensation of fluorescent spillover between channels, which is not a trivial task. Thus, conventional flow cytometric studies have often been limited to a few markers allowing researchers to distinguish cell lineages of interest. This may change as recent progress, in particular the development of spectral flow cytometry, now allows detection of up to 40 markers in flow cytometric analysis.8
Mass cytometry
Mass cytometry, or cytometry by time-of-flight (CyTOF), has emerged as a potent technology enabling simultaneous analysis of ~40 markers in millions of single cells (Fig. 1f). The technology, combining the principle of flow cytometry with mass spectrometry, uses metal isotopes in place of fluorescently labeled antibodies, avoiding the need for compensation of signal spillover. Mass cytometry does however suffer from great cell loss and ideally requires prior knowledge about marker expression. Notably, the technology also allows for high-dimensional imaging.9
Single-cell RNA-sequencing
Single-cell RNA-sequencing (scRNA-seq) can identify and characterize subpopulations of cells in heterogeneous samples based on their full transcriptional profile (Fig. 1g).10 scRNA-seq is especially suited for detection of rare cell types, as these cannot be identified from the average gene expression values obtained by bulk RNA-seq. In addition, scRNA-seq makes it possible to filter out contaminating cells from subsequent analyses of defined subpopulations. A plethora of library preparation protocols11 and sequencing platforms offers large flexibility regarding throughput and sequencing depth. While no prior knowledge is needed about markers, lowly expressed genes are not always detected, and mRNA levels do not necessarily accurately represent the protein expression of a cell. scRNA-seq may however be combined with different technologies for multi-omics single-cell studies, such as index-sorting,12 staining of proteins with barcoded antibodies,13 or combined sequencing of transcriptomes and genomes14 and/or epigenomes (Table 1). scRNA-seq may also be integrated with spatial transcriptomics in order to determine the spatial distribution of cell populations within a tissue and thereby identify local networks of cell communication.15
TCR- and BCR-sequences can be computationally reconstructed directly from scRNA-seq data,16,17,18,19,20,21,22,23 facilitating integrated analysis of antigen specificity and phenotype. This information can also be obtained by commercial platforms, such as the Chromium Single Cell Immune Profiling solution from 10x Genomics.
Celiac disease: a food hypersensitivity disorder with many autoimmune features
Celiac disease (CeD) is a common small intestinal chronic inflammatory disease driven by exposure to cereal gluten proteins of wheat (gliadins and glutenins), barley (hordeins) and rye (secalins).24 The disease varies in severity and clinical presentations with symptoms such as abdominal pain, diarrhea and malnutrition in addition to a long list of extra-intestinal symptoms.25,26 CeD is diagnosed based on clinical, serological and histopathological data from biopsies of the proximal small intestine.27 Patients typically present with blunted or flat intestinal villi, crypt hyperplasia and increased frequencies of lymphocytes in the lamina propria and epithelial layer.28,29,30 CeD can usually be effectively treated with a life-long, strict gluten-free diet.
CeD can be classified as a food hypersensitivity disorder due to the strong immune response toward ingested gluten, but the disease also has autoimmune features such as highly disease-specific autoantibodies, targeted killing of enterocytes, and typical autoimmune genetics with a dominant role of Human Leukocyte Antigen (HLA)-genes.31,32 The disease is virtually only seen in individuals who carry one or two of the HLA-DQ2.5, HLA-DQ2.2 or HLA-DQ8 allotypes.33 These predisposing HLA-DQ variants are also found in many healthy subjects, so “HLA” is a necessary but not sufficient factor for CeD to develop.34 Our current understanding of the mechanisms driving CeD is quite advanced compared to most other autoimmune diseases, and is centered around recognition of gluten-derived peptides on disease-associated HLA-DQ molecules by gluten-specific CD4+ T cells.
Gluten components differ from most other ingested proteins in that they largely resist degradation by digestive enzymes due to their abundance of proline.35 Moreover, gluten peptides are excellent targets for a posttranslational modification, termed deamidation, by the enzyme transglutaminase 2 (TG2).36 TG2 recognizes sequence motifs frequently occurring in gluten-derived peptides to convert specific glutamine residues to glutamate. Such deamidated peptides can be good ligands for disease-associated HLA-DQ molecules, and deamidated gluten peptides (DGP) are recognized by gluten-specific CD4+ T cells of CeD patients when presented on the surface of antigen-presenting cells (APCs).37 In the lamina propria there are gluten-specific effector memory (TEM) cells that drive the development of celiac lesions through secretion of proinflammatory cytokines and likely cross-talk with cytotoxic intraepithelial lymphocytes (IELs).38,39
CeD is also hallmarked by augmented humoral immunity, with increased numbers of lamina propria PCs and enhanced local gut immunoglobulin secretion.40,41,42 Moreover, the patients have disease-specific serum antibodies to gluten43 and TG2.44 Antibodies to gluten is to be expected, but autoantibodies to TG2 is peculiar knowing the food-hypersensitivity nature of CeD. It is intriguing how a foreign antigen like gluten can drive autoimmune responses against TG2,45 especially since there is little evidence for the existence of TG2-specific T cells.38 The dual role of TG2 as the enzyme responsible for deamidation of gluten and for being the autoantigen is hardly coincidental. Production of TG2 autoantibodies conceivably can be explained by gluten-specific CD4+ T cells providing help in a hapten-carrier like fashion to TG2-specific B cells via a formation of gluten:TG2 complexes.46 The gluten-specific T cells can also provide help to gluten-specific B cells (Fig. 2). Of note, antibodies to DGP are particularly specific to the disease, and monitoring of serum antibodies to TG2 and DGP has become an essential part of the diagnostic work-up for CeD.47 The priming of gluten-specific T cells and TG2/DGP-specific B cells likely takes place in organized lymphoid structures like the Peyer’s patches, possibly with involvement of TG2 from shed enterocytes.48 The activated T and B cells would then enter the blood circulation and home to the lamina propria, where they exert effector functions.49
Conceivably, gluten-specific CD4+ T cells can provide help to both transglutaminase 2 (TG2)-specific and gluten-specific B cells. The help to TG2-specific B cells is possible by involvement of TG2:gluten complexes. In a hapten-carrier like fashion, TG2-specific B cells can take up such complexes by BCR-mediated endocytosis, and internalized gluten peptides containing T-cell-relevant gluten epitopes can then be presented to gluten-specific CD4+ T cells bound to the disease-associated HLA-DQ molecules. Gluten-specific B cells also take up gluten peptides, either alone or as part of TG2:gluten complexes, in a similar manner and present them to gluten-specific CD4+ T cells. Antigen presentation to the CD4+ T cell activates both the B cells and T cells, and leads to proliferation of both populations and induction of differentiation pathways.
Detection of antigen-specific cells IN CeD
Gluten-specific CD4+ T cells
Historically, stimulation of gut biopsies with gluten antigen allowed for the isolation of gluten-specific CD4+ T cells, and strikingly these T cells used the disease-associated HLA-DQ variants as restriction molecules.50 The gluten-specific CD4+ T cells are epitope-specific, and many different gluten epitopes exist.51 A few epitopes, termed immunodominant, mount the strongest recall responses and also give responses in most patients.38
Recombinant soluble HLA-DQ molecules with bound gluten peptides represent the ligands for gluten-specific CD4+ T cells, and these molecules when multimerized allow for isolation of the cells at bulk population or single-cell level (Fig. 3a).52,53 Such HLA-DQ:gluten tetramers have been used to identify T cells specific for the immunodominant gluten epitopes DQ2.5-glia-α1a,54,55 DQ2.5-glia-α2,55,56,57, DQ2.5-glia-ω1,54 DQ2.5-glia-ω2,57 DQ2.5-hor358, or DQ8-glia-α153,59 from small intestinal biopsies53,54,56,58,59 and/or peripheral blood54,55,56,57,58,59 of untreated patients, treated patients or treated patients after gluten challenge. Bead enrichment of tetramer-binding cells furthermore enables detection of gluten-specific CD4+ T cells in blood of treated patients, suggesting that tetramer analysis can be a diagnostic tool even for individuals who are already eating a gluten-free diet.60,61,62
a Staining of gluten-specific CD4+ T cells with pMHC class II tetramers. b Staining of transglutaminase 2 (TG2)-specific plasma cells with biotinylated TG2 monomers, visualized by binding of fluorescently labeled streptavidin, or TG2:streptavidin tetramers. c Staining of gluten-specific plasma cells immune-relevant gluten peptides as monomers or tetramers, visualized as in (b).
While many studies of gluten-specific T cells have focused on a particular T-cell epitope, others have used a pool of immunodominant HLA-DQ:gluten tetramers to capture a bigger picture of the T-cell response toward gluten.62,63,64,65 Collectively, the studies have shown that gluten-specific CD4+ cells are selectively present in CeD patients, can readily be detected in the blood and gut after a short-term oral gluten challenge, and decrease in frequency in treated patients.
TG2-specific B-lineage cells
Detection of TG2-specific serum autoantibodies in CeD patients indicates that TG2-specific antibody-secreting cells must be present in these individuals. Using TG2 as bait, TG2-specific PCs have been detected both by immunohistochemistry of duodenal biopsy cryosections and flow cytometry of lamina propria lymphocytes (Fig. 3b). TG2-specific PCs make up on average 10% of all duodenal PCs in untreated CeD.66 While TG2-specific antibodies were frequently identified in the intestine, serum antibody titers did not correlate with the frequency of intestinal TG2-specific PCs, indicating that part of the serum autoantibodies in CeD may be produced outside of the small intestine.67,68 This notion was further corroborated by comparative proteomic analysis of TG2-specific (as well as DGP-specific) antibodies in serum and those produced in mucosal explants, revealing that the antigen-specific serum IgA antibodies were mainly monomers, in contrast to the typical mucosal-derived dimeric form.69
TG2-specific PCs are selectively found in CeD patients.66 Interestingly, they have been detected prior to villous atrophy, and increase in frequency during overt disease.68 This finding fits with autoantibodies commonly being present before the onset of symptoms in CeD.26 Although TG2-specific PCs decrease in frequency on a gluten-free diet,66,67,68,70,71 some cells can be identified even in well-treated patients having been gluten free for years.67,70,71
Little is known about TG2-specific B-cell populations in blood, presumably due to their low abundance. Rare TG2-specific memory B cells have been detected indirectly in CeD patients,70 and we are currently characterizing such cells at a single-cell level.
Gluten-specific B-lineage cells
Gluten-specific intestinal PCs have been identified with a high staining specificity (75–80%) by using tetramerized synthetic gluten peptides as antigen (Fig. 3c).72 Approximately 1% of intestinal IgA+ PCs of untreated CeD patients bound a peptide with the sequence PLQPEQPFP, while on average 0.5% recognized a 33-mer peptide (LQLQPFPQPELPYPQPELPYPQPELPYPQPQPF).72 The frequency of gluten-specific PCs is also dependent on gluten exposure.71 As with TG2-specific PCs, some gluten-specific PCs exist even after years on a gluten-free diet.71
Antigen receptor sequencing
Gluten-specific CD4+ T cells
Biased TCR repertoires against specific pHLA class II complexes have been described in various diseases and can involve preferential V-gene usage, chain pairing, conserved CDR3 motifs, and/or identical TCR sequences across patients.73,74 scAIRR-seq has revealed biased V-gene usage and chain pairing for gluten-specific CD4+ T cells recognizing each immunodominant epitope. In addition, TCRs with similar V-gene usage and sequences have been detected across individuals, terming such TCRs as public receptors.75
The TCR repertoires of gluten-specific CD4+ T cells display extensive use of public TCRs. T cells recognizing the homologous DQ2.5-glia-α1a or DQ2.5-glia-ω1 epitopes have a shared V-gene bias with preferred usage of TRAV35, TRAV4, TRAV12-2, and TRBV29-1 or TRBV20-1 and TRBV5-1.54 DQ2.5-glia-α2-specific T cells preferentially use TRAV26, TRAV4, and TRBV7,56,57,76,77 while T cells specific for the homologous DQ2.5-glia-ω2 epitope preferentially use TRAV4 and TRBV4.57 The DQ2.5-glia-α2-specific TCRs that use the canonical TRAV26-1/TRBV7-2 pair moreover typically have a conserved CDR3β arginine motif.56,57,75,76 T cells specific for the immunodominant barley-derived gluten epitope DQ2.5-hor-3 display a frequent use of TRAV26-1 and TRBV20-1 or TRBV29-1.58 Similar V-gene biases have also been reported for DQ8-restricted gluten epitopes with DQ8-glia-α1-specific T cells preferentially using TRBV9/TRAV26-2 with a conserved CDR3 arginine motif.53,59
CD8+ αβ and γδ IELs
In contrast to the well-characterized gluten-specific CD4+ T cells, the antigen specificities of CD8+ αβ and γδ IELs that increase in frequency in CeD are still elusive.78 A scAIRR-seq study by Han and colleagues using TCRβ or TCRγ sequencing of activated (CD38+ ) and gut-homing (CD103+ ) CD8+ αβ T cells and γδ T cells in blood of CeD patients 6 days after a gluten challenge revealed a high degree of clonal expansion and preferential usage of TRBV7-8, TRBV7-9, and TRBV28 among the CD8+ αβ T cells and TRDV1 (i.e. Vδ1) among the γδ T cells.77 scAIRR-seq of γδ IELs and peripheral blood gut-homing γδ T cells showed bias toward TRGV4 (i.e. Vγ4) and TRDV1 usage in healthy individuals, and demonstrated that the CeD patients displayed a more diverse γδ TCR repertoire with the TRGV4 bias being absent.79 Interestingly, the shift in the γδ TCR repertoire was nearly identical for untreated and treated patients. Unlike γδ T cells, no irreversible change of the TCR repertoire related to CeD has been identified for CD8+ αβ T cells.80 A recent study,80 in contrast to a previous report,77 showed that the CD8+ αβ T cell repertoire is highly polyclonal and diverse in untreated compared to treated CeD and controls.
TG2-specific B-lineage cells
A single-cell study of the BCRs of 60 intestinal TG2-specific PCs indicated a biased V-gene usage and limited number of somatic mutations.66 These characteristic features were further investigated by high-throughput AIRR-seq of the immunoglobulin heavy-chain variable (IGHV) region in gut and blood,70 large-scale scAIRR-seq of intestinal IgA PCs81 and computational BCR sequence reconstruction from scRNA-seq data of intestinal PCs,71 providing ample evidence of preferential V-gene usage and chain pairing, limited somatic mutations and affinity maturation, clonal expansion, low diversity, and skewed isotype usage.
TG2-specific PCs have a particularly prominent bias for the IGHV5-51:IGKV1-5 pair, but there are biases also for several other heavy- and light-chain V-gene pairs such as IGHV3-48:IGLV5-45 and IGHV4-34:IGKV1-39 (Fig. 4a).81 In addition, TG2-specific PCs are skewed toward the IgA isotype,72 in particular IgA1,69,71 compared to the general PC population in CeD and controls.82 The frequencies of somatic mutations correlate for heavy- and light-chain both for TG2-specific PCs and the general PC population, indicating that the low acquisition of mutations is related to the developmental history of the cell.71,81 Typically, many of the mutations retain the physicochemical properties of the amino acid, arguing for limited affinity maturation and conservation of germline-encoded residues.81
a Preferential V-gene pairing of TG2-specific intestinal PCs compared to PCs of unknown specificity. The figure is reproduced from81. b Stereotyped V-gene pairs making up the major part of the repertoire of gluten-specific intestinal PCs. The figure is based on data published in71. Each dot represents a CeD patient, and bars represent the median frequency among the patients.
Gluten-specific B-lineage cells
Gluten-specific B-lineage cells have been less studied than their TG2-specific counterparts, as they are infrequent in comparison. A study of 38 single gluten-specific PCs72 and IGHV bulk sequencing of IgA+ PCs showed repertoire features bearing striking resemblance to TG2-specific PCs, with restricted V-gene usage and limited mutations.83 Recently, paired BCR sequences of 232 gluten-specific intestinal PCs were reconstructed from scRNA-seq data.71 These studies point to a highly stereotypic antibody response to gluten, mostly made up of the chain pairings IGHV3-23/IGLV4-69, IGHV3-15/IGKV4-1, or IGHV3-74/IGKV4-1 (Fig. 4b). Mutational analysis indicated that despite correlation between mutational load in the heavy- and light-chain, their propensity to undergo SHM compared to the general PC population varies with V-gene usage.71 This study also corroborated flow cytometry data72 showing that gluten-specific PCs are enriched for the IgM isotype compared to TG2-specific PCs.
Antigen receptor interactions with antigen
TCR:pHLA-DQ binding
From single CD4+ T cells of CeD patients, several disease-relevant TCR structures in ternary complexes of TCR:pMHC have been resolved, both for TCRs restricted by HLA-DQ2.5,84 HLA-DQ2.285 and HLA-DQ853,59,86. These studies have explained the structural basis for V-gene biases as well as the basis for the conserved CDR3β arginine motif in TCRs using TRAV26-1/TRBV7-2. What remains unexplained at this point is the general deamidation dependence of the gluten-specific CD4+ T cells in CeD.
TG2-specific BCRs/antibodies
Limited mutation of TG2-specific antibodies indicates good affinity in the germline configuration. Indeed, reverting a few TG2-specific hmAbs to their germline sequences has demonstrated that while being reduced, the affinity to TG2 was still relatively high.66 Thus, affinity maturation does not seem to be essential for development of CeD.
CeD patient-derived TG2-specific hmAbs were found to preferentially target the enzyme in its catalytically active conformation, and the antibodies recognize mainly four N-terminal conformational epitopes that are only accessible on soluble TG2 and correlate with IGHV gene usage of the antibodies.66,87,88 Notably, the antibodies did not cross-react with other transglutaminases.87 The structural basis for binding to TG2 has been studied with an epitope 1-specific hmAb using IGHV5-51/IGKV1-5, revealing several residues in both heavy- and light chain to be involved in epitope recognition.81,89
TG2-specific antibodies can be crosslinked by TG2 when expressed as IgD or IgM, but not IgA1 or IgG1, possibly favoring recruitment of PCs from naive or IgM memory cells.66 Interestingly, the functional effects of antibody binding to TG2 vary depending on the epitope, potentially explaining biases toward certain epitopes.90 For instance, antibodies recognizing C-terminal epitopes have been demonstrated to disrupt TG2-mediated crosslinking.88 Furthermore, in vitro engineered B cells specific for N-terminal, but not C-terminal, epitopes could take up TG2-gluten complexes and act as APCs to gluten-specific T cells. Thus, the N-terminal epitope bias could result from B- and T-cell collaboration.88
Gluten-specific BCRs/antibodies
The epitopes recognized by gluten-specific serum antibodies have been characterized by measuring the reactivity to gluten peptide libraries.91,92 Strikingly, the B-cell epitopes are located close to or overlapping with T-cell epitopes within the gluten protein sequences. Of note, DGPs elicit higher antibody reactivity compared to their native counterparts,92,93 indicating a role of deamidation for epitope recognition in CeD.
Expression cloning of 38 gluten-specific hmAbs from single intestinal PCs confirmed that the antibodies typically bound gluten peptides related to T-cell epitopes, and many preferentially bound DGP.72 While the hmAbs were not polyreactive, some exhibited cross-reactivity with different DGPs. Binding of the hmAbs to synthetic gluten peptide was blocked in the presence of patient-derived serum, suggesting recognition of common gluten epitopes.72 A study based on the same hmAb panel demonstrated that the antibodies commonly bind to long deamidated peptide fragments harboring multivalent B-cell epitopes, TG2 recognition sequences, and various T-cell epitopes.94 As for TG2-specific antibodies, there seems to be a correlation between V-gene usage and gluten epitope specificity.72
The stereotyped antibody response toward gluten has made it possible to study the structural basis for antibody recognition of DGP in CeD. Crystal structures for two antibodies using IGHV3-23/IGLV4-69 or IGHV3-15/IGKV4-1 reveal that germline-encoded residues in heavy- and light-chain are involved in binding to the DGP with sequence PLQPEQPFP.83 Some of these residues may be important across a set of gluten-specific antibodies. For instance, DGP-specific antibodies using IGHV3-15/IGKV4-1 or IGHV3-74/IGKV4-1 share a conserved arginine residue in the heavy chain (R55), which is essential for binding of PLQPEQPFP.71,83 R55 is nonpolymorphic in only five IGHV genes, although seven additional genes have one or more R55-containing alleles.95 Importance of conserved germline residues could argue for a role of polymorphisms in the antibody response in CeD. As a proof of concept, it was demonstrated that a patient-derived gluten-specific antibody using the R55-containing IGHV4-4*07 allele paired with IGKV4-1 could not bind to DGP when mutating R55 to the allelic variant E55.71
Lineage tracing
For a long time, it was unknown whether the circulating gluten-specific T cells were representative of the larger population of CD4+ T cells residing in the lamina propria. Bulk and scAIRR-seq of tetramer-sorted CD4+ T cells in CeD revealed the two compartments to be composed of nearly identical T-cell clonotypes both in untreated and treated CeD, indicating a common origin and trafficking of clones between blood and gut.63
Disease-relevant T-cell clonotypes in CeD are not only shared between gut and blood, but also at different time points. This has been shown for gluten-specific CD4+ T cells63 and also for CD103+ γδ and CD8+ T cells.96 Further, by comparing single T cells from intestinal biopsies taken years apart, gluten-specific CD4+ T-cell clonotypes were demonstrated to persist for decades in CeD patients on a gluten-free diet.63 Moreover, the recall response during gluten challenge was dominated by preexisting clonotypes. Thus, the disease-specific TCR repertoire remains stable over years in treated patients. While long-lived intestinal memory T cells exist,97,98 occasional trace amounts of gluten likely contribute to these stable T-cell responses.
To date, only two studies have traced the evolution of intestinal CeD-specific PC lineages at a single-cell level, revealing limited affinity maturation and frequent expansion of cells with identical BCRs within each clone.71,81 Interestingly, PCs expressing different isotypes and longevity-associated phenotypes were found within individual clones, indicating ongoing immune responses toward TG2 and DGP involving isotype class switching and potentially reactivation of memory B cells.71 Combined lineage tracing of PCs and memory B cells could be an important tool to answer whether reactivation of memory B cells contributes to the pool of TG2- or DGP-specific PCs. The only published study to date using this approach was at the bulk level, indicating a limited overlap between TG2-specific PCs and circulating memory B cells.70 Clonal overlaps have also been shown by a combination of bulk IGHV sequencing of intestinal PCs and proteomic analysis of the heavy-chain CDR3 region of serum antibodies.69 Detailed studies at single-cell resolution, preferably from multiple time points and tissues, on and off a gluten-free diet or during gluten challenge, are warranted.
Single-cell phenotyping of adaptive immune cells in celiac disease
Several studies have aimed to phenotype adaptive immune responses in CeD at a single-cell resolution to better understand their function and identify potential therapeutic targets. In recent years, powerful single-cell “omics” studies have emerged, shedding new light on T- and B-cell responses at the protein and mRNA levels (Table 2).
Assessing phenotypes by immunohistochemistry and flow cytometry
T-cell populations
Altered compositions of T-cell subtypes have been identified in the intestine and blood of CeD patients compared to controls.99,100,101 An immunohistochemistry-based study demonstrated that intestinal CD4+ T cells lack expression of the proliferation marker Ki-67, but express the activation marker CD25 upon gluten exposure.102
Gluten-specific CD4+ cells
Tetramer visualization of gluten-specific CD4+ T cells has allowed further dissection of the phenotypes of disease-relevant T cells in CeD. Circulating DQ2.5-glia-α1a-specific T cells of CeD patients undergoing a gluten challenge expressed integrin β7, indicating that they are gut-homing.55 Further, phenotyping of gluten-specific CD4+ T subsets in blood by tetramer staining of naive, TEM and central memory (TCM) cells revealed increased frequencies of gluten-specific TEM cells, in particular gut-homing cells, in both untreated and treated CeD compared to controls.61 It was reported that a large proportion of circulating gluten-specific CD4+ T cells are Tregs, based on their expression of FOXP3 and CD39, and a paradoxical higher number of these cells in CeD patients was explained by the cells having impaired suppressive functions.103 Others found increased frequencies of gluten-specific TH17 cells in CeD.104 While very few gluten-reactive TCM or TEM cells can be detected in controls, gluten-specific naive T cells are found in controls as well as in CeD patients.61 Functional studies of HLA-DQ:gluten tetramer-positive cells later revealed that healthy HLA-DQ2.5+ controls lack gluten-specific regulatory and memory T cells.105
IELs
While their antigen specificity is unknown, a fraction of IELs express Ki-67 after gluten exposure, indicating proliferation.102 Activated (CD38+ ), gut-homing (CD103+ ) γδ and CD8+ αβ T cells have also been detected in blood following gluten challenge.77 CD8+ αβ T cells constitutively express activating natural killer receptors,106,107 with the highest expression of the activating receptor NKG2D found in the intestine of untreated compared to treated CeD patients.108 While most small intestinal γδ IELs express the activating natural cytotoxicity receptors NKp46 and/or NKp44, their expression is significantly reduced in CeD patients, even after years on a gluten-free diet.109 Furthermore, patients on a gluten-free diet have been shown to have higher frequencies of CD8+ γδ IELs expressing the inhibitory receptor NKG2A and intracellular TGF-β1 compared to untreated patients.110 While CD8+ αβ IEL frequencies normalize on a gluten-free diet, this is not the case for γδ IELs.111
The dynamic regulation of different T-cell populations during gluten challenge is under some debate. While Han et al. and others have reported increased frequencies of both gluten-specific CD4+ T cells, γδ T cells, and CD8+ αβ T cells in blood after gluten challenge,77,112 it was recently reported that not all patients respond with an increase of all three T-cell subsets in blood, although all patients with an increase of γδ and/or CD8+ αβ T cells also showed elevated gluten-specific CD4+ T-cell frequencies.96 The discrepancies between studies could potentially be ascribed to differences in gluten challenge and laboratory protocols, and illustrates a need for standardized challenge protocols.113,114
Intestinal PCs
The large population of intestinal antibody-secreting cells in CeD patients and controls are mainly end-differentiated PCs rather than proliferating plasmablasts, as they express CD138 and lack CD20 or intracellular staining for Ki-67.4,66 PCs of the IgA and IgM isotypes express a functional BCR,4,115 and at least a proportion of intestinal PCs retain surface expression of HLA class II4,66,116 and costimulatory116 molecules.
Intestinal PCs can, despite previous beliefs, survive for years. Notably, retrospective carbon-14 dating of bulk-sorted populations based on the cell surface proteins CD19 and CD45 has revealed three populations of short-, intermediate-, and long-lived PCs with a median age of 0, 11, and 22 years, respectively.117 Along the same theme, it was recently demonstrated that most PCs in untreated CeD, in particular TG2- or DGP-specific PCs, have a short- or intermediate-lived phenotype, possibly reflecting the large expansion of PCs in the celiac lesion.71 This was true also for patients staying gluten free for a few years, while patients treated for more than a decade mainly had long- and intermediate-lived TG2- or DGP-specific PCs. The high frequency of short-lived TG2- or DGP-specific PCs in patients treated for several years was surprising, and possibly indicates that these patients unknowingly may have been exposed to minute amounts of gluten.118
Memory B cells
No study has characterized the phenotype of memory B cells in CeD.
Regulatory B cells
Regulatory B cells (Bregs) have been implicated in various autoimmune conditions.119 Although TG2- or gluten-specific Bregs have yet to be identified in CeD, the general population of peripheral blood CD24hi CD38hi Bregs has been shown to be larger in CeD patients compared to controls or patients with the autoimmune disease Hashimoto’s thyroiditis.120 The same authors have proposed that these Bregs may be functionally defective in CeD based on their limited ability to produce interleukin (IL)-10 upon in vitro stimulation with CpG.
Phenotyping by mass cytometry
A study by van Unen et al.121 used mass cytometry to analyze the overall composition of innate and adaptive immune cells in intestinal biopsies and paired PBMCs from healthy individuals and patients with inflammatory intestinal diseases, including CeD. The study confirmed previous reports of an increased γδ IEL population in CeD, and further identified differences in innate lymphocyte populations.
T cells
A limited number of mass cytometry studies have focused on the T-cell response in CeD. This technology was used in a study on activated and gut-homing γδ and CD8 + αβ T cells after gluten challenge.77 A later study combined HLA tetramer staining with mass cytometry to phenotype gluten-specific CD4+ T cells.122 Interestingly, this study revealed that gluten-specific CD4+ T cells have a distinct phenotype with upregulation of activation markers CD38, CD161, CD28, HLA-DR and OX40 as well as CXCR3, CD39, and PD-1 both in the gut and blood (Fig. 5). Remarkably, the same CD4+ T-cell phenotype was seen in patients with systemic sclerosis or systemic lupus erythematosus, suggesting that disease-driving CD4+ T cells in autoimmune conditions with unknown target antigens could be identified based on their phenotype.
B-lineage cells
To date, no studies using mass cytometry to dissect B-cell responses in CeD have been published. Although the study by van Unen et al. contained some B-lineage cells, it lacked markers to dissect the B-lineage response. While studies of antigen-specific B-lineage cells in CeD are warranted, characterization of B-cell subsets and their marker expression in the healthy intestine and blood may serve as a reference.123 Mass cytometry has already been used in combination with staining of antigen-specific cells to profile rotavirus-specific intestinal and circulating B-lineage cells.123
Single-cell RNA-sequencing
scRNA-seq has the potential to build on previous bulk RNA-seq studies of CeD122,124,125,126,127,128 by dissecting the cellular composition of celiac lesions, identifying rare cell types potentially involved in disease pathology, and excluding contaminating cells from detailed analyses of specific cell populations. With the goal of characterizing the composition of all immune cells in the celiac lesion, Atlasy and colleagues sequenced immune cells from duodenal biopsies from celiac patients and controls.129 Corroborating previous findings, the authors reported increased numbers of total lamina propria immune cells and CD3+ IELs in untreated CeD patients compared to controls.
T cells
scRNA-seq studies of T cells with known antigen specificity in CeD are currently lacking; the only RNA-seq data available for gluten-specific T cells is from bulk populations, largely agreeing with mass cytometry data.122 While no information on antigen specificity is included, the preprint by Atlasy et al. found the majority of CD4+ T cells to differ transcriptionally in CeD patients and controls. Furthermore, most of the epithelial and lamina propria CD8+ T cells had an activated phenotype in CeD compared to controls, with expression of cytotoxic activity-associated genes such as KLRC2 and KLRD1.
B-lineage cells
A recent scRNA-seq study of intestinal PCs in CeD and controls largely corroborated bulk RNA-seq data,124 and revealed that TG2- and DGP-specific PCs differ to some extent from PCs of unknown specificity.71,124 At least a subset of the PCs, regardless of antigen specificity, expressed low levels of mRNA transcripts for HLA class II and costimulatory molecules. Interestingly, the general intestinal PC population in CeD patients may differ transcriptionally from controls.71,124 Differences between PCs in untreated and treated CeD and controls may indicate potentially pathogenic roles of PCs, including non-disease-specific ones, in CeD.
PCs typically have been defined as CD27high, in addition to high expression of CD38 or CD138. Likewise, CD27 has been widely used as a memory B cell marker. These definitions are however changing, as CD27−/dim B-cell subsets are gaining more interest,130,131 especially in the context of autoimmune diseases. Indeed, our RNA-seq studies revealed that CD27 expression is lower in TG2- or DGP-specific PCs compared to other PCs, as well as in PCs of CeD patients compared to controls. Enrichment of CD27−/dim TG2-specific PCs and circulating memory B cells has also been observed at the protein level (our unpublished results).
T-cell functions in CeD
Helper cell functions
The pile of evidence resulting from extensive CeD research points to gluten-specific CD4+ T cells as key players in CeD pathogenesis by providing help to B cells for antibody production132 and by supporting cytotoxic IELs that kill enterocytes.38 Bulk RNA-seq data of HLA-DQ:gluten tetramer-staining cells of the gut lesion revealed expression of IL21 and CXCL13, cytokines hallmarking follicular helper cells providing B-cell help,133 yet a lack of CXCR5 expression speaks against a germinal center function of these cells.122 Of note, the interaction of T cells with B cells does not only affect the B-cell side resulting in antibody production, it also leads to T-cell activation and clonal expansion. Studies of transfectants where CeD patient-derived TCRs and BCRs were introduced demonstrated that gluten-specific T cells can be activated by TG2-specific B cells in the presence of TG2:gluten complexes.66
Regulatory T cell function
Tregs are a subset of CD4+ T cells, important for maintaining peripheral tolerance. It has therefore been speculated that in the healthy state a response toward gluten is kept in check by Treg activity.134,135,136 While expression of the transcription factor FOXP3 is a hallmark of regulatory T cells in mice, robust markers for human Tregs are lacking. Tregs in humans are often characterized as FOXP3+ CD25+ CD127low, albeit FOXP3 can be expressed by activated human CD4+ T cells without regulatory functions.137 A report of high frequencies of gluten-specific CD4+ memory T cells in CeD expressing typical Treg markers following stimulation of blood samples with gluten is puzzling. These cells displayed an impaired suppressive function after in vitro expansion, suggesting that Treg dysfunction may play a role in CeD.103 The results of the study contrast extensive phenotyping of gluten-specific T cells by mass cytometry, which revealed that while some gluten-specific T cells express FOXP3, these cells do not express CD25.122 This discrepancy between studies could be explained by the finding that in vitro stimulation of gluten-specific FOXP3+ CD25- T cells induce CD25 in these cells.122
Healthy HLA-DQ2.5 expressing individuals have rare CD4+ T cells in blood that stain with HLA-DQ:gluten tetramers. These cells do not express FOXP3 or CD25 suggesting that they are not Tregs.105
Cytotoxic functions and other functions by IELs
CD8+ αβ IELs
IELs normally contribute to immune protection of the gut by preventing pathogen entry into the epithelium by maintaining barrier integrity. In CeD, however, the expanded population of CD8+ αβ IELs is thought to be responsible for killing of epithelial cells, contributing to the formation of the disease lesion. The cytokine interleukin IL-15 is considered particularly important for the expansion of the CD8+ αβ IELs138,139, yet other cytokines, like IL-2 and IL-21, are likely also implicated.140,141
It has been suggested that CD8+ IELs in CeD are activated in a TCR-independent fashion by binding of ligands to activating natural killer receptors.38 This hypothesis is supported by the observed polyclonal TCR repertoire and lack of V-gene bias of CD8+ IELs in CeD,80 shift in expression toward activating natural killer receptors in untreated CeD,110 and ability to respond to stress signals via activating natural killer receptors independent of the TCR specificity.136,138 Thus, it is probable that CD8+ IELs sense intestinal stress and inflammation, become licensed to kill enterocytes by help of pro-inflammatory CD4+ T cells, and upregulate their cytolytic activity.
γδ IELs
γδ IELs play an important role in immune surveillance of the intestinal epithelium.142 Their role in CeD pathogenesis is poorly understood. IELs are likely recruited to the intestinal epithelium in CeD as a response to stress or damaged epithelial cells. It has been suggested that a subset of γδ IELs, characterized by expression of CD8 and NKG2A, may play a regulatory role in CeD, based on their ability to limit the cytotoxic potential of IL-15-induced CD8+ αβ IELs.110 Further it was demonstrated that gluten-induced inflammation leads to depletion of naturally occurring Vγ4+/Vδ1+ IELs with innate cytolytic properties and specificity for enterocyte-expressed butyrophilin-like (BTNL) molecules BTNL3/BTNL8.109 Reduced enterocyte expression of BTNL8 and loss of Vγ4+/Vδ1+ IELs was found to be accompanied by the expansion of gluten-sensitive, interferon-γ-producing Vδ1+ IELs that do not recognize BTNL3/BTNL8 and which potentially have a pathogenic role.
B-cell and plasma-cell functions in CeD
Antibody secretion
While autoantibodies may have a clear pathogenic role in some autoimmune diseases, it is uncertain whether antibodies specific to TG2 or gluten are pathogenic as circulating immunoglobulins.143,144 The finding that gluten-specific PCs are enriched for IgM-expressing cells compared to TG2-specific PCs71,72 could have implications for the possible pathogenicity of these antibodies in CeD, as IgM antibodies can activate complement through the classical pathway.145
Antigen presentation
While circulating immunoglobulin may not be pathogenic in CeD, immunoglobulin as BCR on antigen-presenting B cells likely plays an important pathogenic role. In CeD the interaction between antigen-presenting B cells and T cells probably happens in organized lymphoid structures, like Peyer’s patches, isolated lymphoid follicles or mesenteric lymph nodes. The rapid decline of serum antibody levels when patients commence a gluten-free diet146,147 and the limited degree of SHM in TG2-specific66,70,71,81 and DGP-specific71,72,83 PCs at the same time argues for short-lived germinal center or extrafollicular responses.148 Whether there is also interaction of B-lineage cells with T cells in the lamina propria is still an open question. At this site, PCs dominate and there are very few B cells present.149 PCs have been thought to downregulate surface BCR and HLA class II expression, and the cells have hence not been considered as APCs. Importantly, however, IgA- and IgM-expressing PCs, by contrast to IgG-expressing PCs, retain functional cell-surface BCR.4,115 A low/medium expression of HLA class II molecules detected both on the protein and mRNA level in a proportion of intestinal PCs,71,116,124 and an expression of costimulatory molecules116 thus raise the possibility that the expanded lamina propria PC population may act as APCs in CeD. By use of pHLA-specific hmAbs it was demonstrated that PCs are the dominating cell type presenting gluten peptides on their surface.116 Thus, intestinal PCs have the machinery needed to present relevant T-cell epitopes on HLA-DQ molecules on their surface. Importantly, actual T-cell recognition of and activation by PC-displayed gluten peptides is yet to be demonstrated.
Cytokine and chemokine secretion
It is unknown whether B-lineage cells secrete molecules contributing to the development of celiac lesions, as the most abundant B-lineage cells in CeD are PCs, whose functions beyond antibody secretion are not well studied. Recent literature contains some reports of cytokine and chemokine secretion by PCs, particularly by regulatory PCs.119 While there is no evidence of intestinal regulatory PCs in CeD by scRNA-seq, at least a subset of intestinal PCs, regardless of specificity, produce mRNA for IL15 and IL16,71,124 and small populations of PCs express high levels of CCL3 and CCL4.71 This is interesting, given the assumed vital role of IL-15 in CeD pathogenesis,138,139 and as IL-16 attracts and activates CD4+ T cells.150 It is however not known if mRNA expression of these cytokines translates to expression of the secreted cytokines; translation of IL15 is for instance under extensive regulation,151,152,153 and isoforms with different functions and subcellular localization exist.153,154
Concluding remarks and future directions
In this review we have discussed how single-cell approaches have provided invaluable knowledge about the specificity, binding mode, phenotype, and function of antigen-specific adaptive immune cells in the well-characterized disease CeD. As community efforts such as the Human Cell Atlas155 have gained substantial interest in recent years, we expect that new mass cytometry and scRNA-seq studies will continue to build the basis for detailed maps of the phenotypes of T- and B-cell populations found in various tissues of autoimmune patients and healthy individuals. Single-cell multi-omics studies will likely contribute to new knowledge about CeD pathogenesis and may uncover novel potential therapeutic targets. Of note, integration of spatial transcriptomics data with scRNA-seq and protein expression could reveal networks of communicating immune cells and their interactions with epithelial and stromal cells in the celiac lesion, improving our understanding of the mechanisms underlying CeD pathology.
The lack of knowledge of the disease-driving antigens remains a major hurdle for further progress in understanding the pathogenesis of many autoimmune diseases. Single-cell analysis may well provide help to overcome this hurdle. The finding of a rare phenotypic profile in CD4+ T cells that is shared across diseases may offer an avenue to define the disease-driving T cells and the disease-driving antigen in diseases where this is unknown.122 Further, single-cell analysis allows to define clonally expanded populations of T and B cells. In the lesion of the disease, culprit T and B cells are likely to belong to the clonally expanded cells. If there among clonally expanded cells are stereotypic patterns shared across patients, the likelihood that these cells are disease-relevant would increase. For instance, if the autoantigen of celiac disease was unknown, single-cell sequencing of PCs in the celiac lesions probably would have identified clonally expanded IGHV5-51/IGKV1-5 PCs. Expression cloning of such IGHV5-51/IGKV1-5 monoclonal antibodies, with subsequent identification of their antigen ligands, would then identify TG2 as the target antigen. If also clonally expanded T cells exist, showing a productive interaction for transfectants armed with TCRs and BCRs of the clonally expanded cells would represent a further step toward identifying the disease-driving antigen. Further certainty would come if the CD4+ T cells use disease-associated HLA allotypes as their restriction element. So far, we lack examples that demonstrate the success of such approaches. Notwithstanding, we are sure we have just seen the beginning of single-cell analysis of autoimmune conditions. Exciting years lie ahead of us.
References
Franz, B., May, K. F. Jr., Dranoff, G. & Wucherpfennig, K. Ex vivo characterization and isolation of rare memory B cells with antigen tetramers. Blood 118, 348–357 (2011).
Altman, J. D. et al. Phenotypic analysis of antigen-specific T lymphocytes. Science 274, 94–96 (1996).
Crawford, F., Kozono, H., White, J., Marrack, P. & Kappler, J. Detection of antigen-specific T cells with multivalent soluble class II MHC covalent peptide complexes. Immunity 8, 675–682 (1998).
Di Niro, R. et al. Rapid generation of rotavirus-specific human monoclonal antibodies from small-intestinal mucosa. J. Immunol. 185, 5377–5383 (2010).
Hoehn, K. B. et al. Repertoire-wide phylogenetic models of B cell molecular evolution reveal evolutionary signatures of aging and vaccination. Proc. Natl Acad. Sci. USA 116, 22664–22672 (2019).
Stern, J. N. H. et al. B cells populating the multiple sclerosis brain mature in the draining cervical lymph nodes. Sci. Transl. Med. 6, 248ra107–248ra107 (2014).
McKinnon, K. M. Flow cytometry: an overview. Curr. Protoc. Immunol. 120, 5.1.1–5.1.11 (2018).
Bonilla, D. L., Reinin, G. & Chua, E. Full spectrum flow cytometry as a powerful technology for cancer immunotherapy research. Front. Mol. Biosci. 7, (2021).
Hartmann, F. J. & Bendall, S. C. Immune monitoring using mass cytometry and related high-dimensional imaging approaches. Nat. Rev. Rheumatol. 16, 87–99 (2020).
Stubbington, M. J. T., Rozenblatt-Rosen, O., Regev, A. & Teichmann, S. A. Single-cell transcriptomics to explore the immune system in health and disease. Science 358, 58–63 (2017).
Svensson, V. et al. Power analysis of single-cell RNA-sequencing experiments. Nat. Methods 14, 381 (2017).
Wilson, N. K. et al. Combined single-cell functional and gene expression analysis resolves heterogeneity within stem cell populations. Cell. Stem Cell 16, 712–724 (2015).
Stoeckius, M. et al. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 14, 865–868 (2017).
Macaulay, I. C. et al. G&T-seq: parallel sequencing of single-cell genomes and transcriptomes. Nat. Methods 12, 519–522 (2015).
Longo, S. K., Guo, M. G., Ji, A. L. & Khavari, P. A. Integrating single-cell and spatial transcriptomics to elucidate intercellular tissue dynamics. Nat. Rev. Genet. (2021).
Afik, S. et al. Targeted reconstruction of T cell receptor sequence from single cell RNA-seq links CDR3 length to T cell differentiation state. Nucleic Acids Res. 45, e148 (2017).
Canzar, S., Neu, K. E., Tang, Q., Wilson, P. C. & Khan, A. A. BASIC: BCR assembly from single cells. Bioinformatics 33, 425–427 (2017).
Eltahla, A. A. et al. Linking the T cell receptor to the single cell transcriptome in antigen‐specific human T cells. Immunol. Cell Biol. 94, 604–611 (2016).
Stubbington, M. J. T. et al. T cell fate and clonality inference from single-cell transcriptomes. Nat. Methods 13, 329–332 (2016).
Upadhyay, A. A. et al. BALDR: a computational pipeline for paired heavy and light chain immunoglobulin reconstruction in single-cell RNA-seq data. Genome Med. 10, 20 (2018).
Lindeman, I. et al. BraCeR: B-cell-receptor reconstruction and clonality inference from single-cell RNA-seq. Nat. Methods 15, 563–565 (2018).
Rizzetto, S. et al. B-cell receptor reconstruction from single-cell RNA-seq with VDJPuzzle. Bioinformatics 34, 2846–2847 (2018).
Afik, S., Raulet, G. & Yosef, N. Reconstructing B-cell receptor sequences from short-read single-cell RNA sequencing with BRAPeS. Life Sci. Alliance 2, e201900371 (2019).
Shewry, P. What Is gluten-why is it special? Front. Nutr. 6, 101 (2019).
Ferguson, A., Arranz, E. & O’Mahony, S. Clinical and pathological spectrum of coeliac disease–active, silent, latent, potential. Gut 34, 150–151 (1993).
Green, P. H. The many faces of celiac disease: clinical presentation of celiac disease in the adult population. Gastroenterology 128, S74–S78 (2005).
Al-Toma, A. et al. European Society for the Study of Coeliac Disease (ESsCD) guideline for coeliac disease and other gluten-related disorders. U. Eur. Gastroenterol. J. 7, 583–613 (2019).
Sakula, J. & Shiner, M. Coeliac disease with atrophy of the small-intestine mucosa. Lancet 273, 876–877 (1957).
Ferguson, A. & Murray, D. Quantitation of intraepithelial lymphocytes in human jejunum. Gut 12, 988–994 (1971).
Marsh, M. N. Studies of intestinal lymphoid tissue. XI–The immunopathology of cell-mediated reactions in gluten sensitivity and other enteropathies. Scanning Microsc. 2, 1663–1684 (1988).
Sollid, L. M. & Jabri, B. Triggers and drivers of autoimmunity: lessons from coeliac disease. Nat. Rev. Immunol. 13, 294–302 (2013).
Sollid, L. M. & Jabri, B. Is celiac disease an autoimmune disorder? Curr. Opin. Immunol. 17, 595–600 (2005).
Sollid, L. M. The roles of MHC class II genes and post-translational modification in celiac disease. Immunogenetics 69, 605–616 (2017).
Abadie, V., Sollid, L. M., Barreiro, L. B. & Jabri, B. Integration of genetic and immunological insights into a model of celiac disease pathogenesis. Annu. Rev. Immunol. 29, 493–525 (2011).
Shan, L. et al. Structural basis for gluten intolerance in celiac sprue. Science 297, 2275–2279 (2002).
Jabri, B. & Sollid, L. M. Mechanisms of disease: immunopathogenesis of celiac disease. Nat. Clin. Pract. Gastroenterol. Hepatol. 3, 516–525 (2006).
Molberg, Ø. et al. Tissue transglutaminase selectively modifies gliadin peptides that are recognized by gut-derived T cells in celiac disease. Nat. Med. 4, 713–717 (1998).
Jabri, B. & Sollid, L. M. T cells in celiac disease. J. Immunol. 198, 3005–3014 (2017).
Nilsen, E. M. et al. Gluten induces an intestinal cytokine response strongly dominated by interferon gamma in patients with celiac disease. Gastroenterology 115, 551–563 (1998).
Douglas, A. P., Crabbé, P. A. & Hobbs, J. R. Immunochemical studies of the serum, intestinal secretions and intestional mucosa in patients with adult celiac disease and other forms of the celiac syndrome. Gastroenterology 59, 414–425 (1970).
Soltoft, J. Immunoglobulin-containing cells in non-tropical sprue. Clin. Exp. Immunol. 6, 413–420 (1970).
Lancaster-Smith, M., Kumar, P., Marks, R., Clark, M. L. & Dawson, A. M. Jejunal mucosal immunoglobulin-containing cells and jejunal fluid immunoglobulins in adult coeliac disease and dermatitis herpetiformis. Gut 15, 371–376 (1974).
Scott, H., Fausa, O., Ek, J. & Brandtzaeg, P. Immune response patterns in coeliac disease. Serum antibodies to dietary antigens measured by an enzyme linked immunosorbent assay (ELISA). Clin. Exp. Immunol. 57, 25–32 (1984).
Dieterich, W. et al. Identification of tissue transglutaminase as the autoantigen of celiac disease. Nat. Med. 3, 797–801 (1997).
Iversen, R. & Sollid, L. M. Autoimmunity provoked by foreign antigens. Science 368, 132–133 (2020).
Sollid, L. M., Molberg, Ø., McAdam, S. & Lundin, K. E. Autoantibodies in coeliac disease: tissue transglutaminase–guilt by association? Gut 41, 851–852 (1997).
Caja, S., Maki, M., Kaukinen, K. & Lindfors, K. Antibodies in celiac disease: implications beyond diagnostics. Cell. Mol. Immunol. 8, 103–109 (2011).
Iversen, R. et al. Evidence that pathogenic transglutaminase 2 in celiac disease derives from enterocytes. Gastroenterology 159, 788–790 (2020).
Macpherson, A. J., McCoy, K. D., Johansen, F. E. & Brandtzaeg, P. The immune geography of IgA induction and function. Mucosal Immunol. 1, 11–22 (2008).
Lundin, K. E. et al. Gliadin-specific, HLA-DQ(α1*0501,β1*0201) restricted T cells isolated from the small intestinal mucosa of celiac disease patients. J. Exp. Med. 178, 187–196 (1993).
Sollid, L. M. et al. Update 2020: nomenclature and listing of celiac disease-relevant gluten epitopes recognized by CD4+ T cells. Immunogenetics 72, 85–88 (2020).
Quarsten, H. et al. Staining of celiac disease-relevant T cells by peptide-DQ2 multimers. J. Immunol. 167, 4861–4868 (2001).
Petersen, J. et al. Determinants of gliadin-specific T cell selection in celiac disease. J. Immunol. 194, 6112–6122 (2015).
Dahal-Koirala, S. et al. Discriminative T-cell receptor recognition of highly homologous HLA-DQ2-bound gluten epitopes. J. Biol. Chem. 294, 941–952 (2019).
Ráki, M. et al. Tetramer visualization of gut-homing gluten-specific T cells in the peripheral blood of celiac disease patients. Proc. Natl Acad. Sci. USA 104, 2831–2836 (2007).
Qiao, S. W., Christophersen, A., Lundin, K. E. & Sollid, L. M. Biased usage and preferred pairing of α- and β-chains of TCRs specific for an immunodominant gluten epitope in coeliac disease. Int. Immunol. 26, 13–19 (2014).
Dahal-Koirala, S. et al. TCR sequencing of single cells reactive to DQ2.5-glia-α2 and DQ2.5-glia-ω2 reveals clonal expansion and epitope-specific V-gene usage. Mucosal Immunol. 9, 587–596 (2016).
Dahal-Koirala, S., Neumann, R. S., Jahnsen, J., Lundin, K. E. A. & Sollid, L. M. On the immune response to barley in celiac disease: biased and public T-cell receptor usage to a barley unique and immunodominant gluten epitope. Eur. J. Immunol. 50, 256–269 (2020).
Broughton, S. E. et al. Biased T cell receptor usage directed against human leukocyte antigen DQ8-restricted gliadin peptides is associated with celiac disease. Immunity 37, 611–621 (2012).
Brottveit, M. et al. Assessing possible celiac disease by an HLA-DQ2-gliadin tetramer test. Am. J. Gastroenterol. 106, 1318–1324 (2011).
Christophersen, A. et al. Tetramer-visualized gluten-specific CD4+ T cells in blood as a potential diagnostic marker for coeliac disease without oral gluten challenge. U. Eur. Gastroenterol. J. 2, 268–278 (2014).
Sarna, V. K. et al. HLA-DQ-gluten tetramer blood test accurately identifies patients with and without celiac disease in absence of gluten consumption. Gastroenterology 154, 886–896.e886 (2018).
Risnes, L. F. et al. Disease-driving CD4+ T cell clonotypes persist for decades in celiac disease. J. Clin. Invest. 128, 2642–2650 (2018).
Zühlke, S. et al. CD38 expression on gluten-specific T cells is a robust marker of gluten re-exposure in coeliac disease. U. Eur. Gastroenterol. J. 7, 1337–1344 (2019).
Sarna, V. K. et al. HLA-DQ:gluten tetramer test in blood gives better detection of coeliac patients than biopsy after 14-day gluten challenge. Gut 67, 1606–1613 (2018).
Di Niro, R. et al. High abundance of plasma cells secreting transglutaminase 2-specific IgA autoantibodies with limited somatic hypermutation in celiac disease intestinal lesions. Nat. Med. 18, 441–445 (2012).
Di Niro, R. et al. Responsive population dynamics and wide seeding into the duodenal lamina propria of transglutaminase-2-specific plasma cells in celiac disease. Mucosal Immunol. 9, 254–264 (2016).
Hietikko, M. et al. Small-intestinal TG2-specific plasma cells at different stages of coeliac disease. BMC Immunol. 19, 36 (2018).
Iversen, R. et al. Strong clonal relatedness between serum and gut IgA despite different plasma cell origins. Cell. Rep. 20, 2357–2367 (2017).
Snir, O. et al. Analysis of celiac disease autoreactive gut plasma cells and their corresponding memory compartment in peripheral blood using high-throughput sequencing. J. Immunol. 194, 5703–5712 (2015).
Lindeman, I. et al. Longevity, clonal relationship, and transcriptional program of celiac disease-specific plasma cells. J. Exp. Med. 218, e20200852 (2021).
Steinsbø, Ø. et al. Restricted VH/VL usage and limited mutations in gluten-specific IgA of coeliac disease lesion plasma cells. Nat. Commun. 5, 4041–4041 (2014).
Miles, J. J., Douek, D. C. & Price, D. A. Bias in the αβ T-cell repertoire: implications for disease pathogenesis and vaccination. Immunol. Cell. Biol. 89, 375–387 (2011).
Turner, S. J., Doherty, P. C., McCluskey, J. & Rossjohn, J. Structural determinants of T-cell receptor bias in immunity. Nat. Rev. Immunol. 6, 883–894 (2006).
Dahal-Koirala, S. et al. Comprehensive analysis of CDR3 sequences in gluten-specific T-cell receptors reveals a dominant R-motif and several new minor motifs. Front. Immunol. 12, 639672–639672 (2021).
Qiao, S. W. et al. Posttranslational modification of gluten shapes TCR usage in celiac disease. J. Immunol. 187, 3064–3071 (2011).
Han, A. et al. Dietary gluten triggers concomitant activation of CD4+ and CD8+ αβ T cells and γδ T cells in celiac disease. Proc. Natl Acad. Sci. USA 110, 13073–13078 (2013).
Mayassi, T. & Jabri, B. Human intraepithelial lymphocytes. Mucosal Immunol. 11, 1281–1289 (2018).
Eggesbø, L. M. et al. Single-cell TCR sequencing of gut intraepithelial γδ T cells reveals a vast and diverse repertoire in celiac disease. Mucosal Immunol. 13, 313–321 (2020).
Eggesbø, L. M. et al. Single-cell TCR repertoire analysis reveals highly polyclonal composition of human intraepithelial CD8+ αβ T lymphocytes in untreated celiac disease. Eur. J. Immunol. 51, 1542–1545 (2021).
Roy, B. et al. High-throughput single-cell analysis of B cell receptor usage among autoantigen-specific plasma cells in celiac disease. J. Immunol. 199, 782–791 (2017).
Chiba, M., Ohta, H., Yagisawa, H. & Masamune, O. IgA1 & IgA2 distribution in the intestine. Gastroenterol. Jpn. 22, 18–23 (1987).
Snir, O. et al. Stereotyped antibody responses target posttranslationally modified gluten in celiac disease. JCI Insight 2, e93961 (2017).
Petersen, J. et al. T-cell receptor recognition of HLA-DQ2-gliadin complexes associated with celiac disease. Nat. Struct. Mol. Biol. 21, 480–488 (2014).
Ting, Y. T. et al. A molecular basis for the T cell response in HLA-DQ2.2 mediated celiac disease. Proc. Natl Acad. Sci. USA 117, 3063–3073 (2020).
Petersen, J. et al. Diverse T cell receptor gene usage in HLA-DQ8-associated celiac disease converges into a consensus binding solution. Structure 24, 1643–1657 (2016).
Iversen, R. et al. Transglutaminase 2-specific autoantibodies in celiac disease target clustered, N-terminal epitopes not displayed on the surface of cells. J. Immunol. 190, 5981–5991 (2013).
Iversen, R. et al. Efficient T cell-B cell collaboration guides autoantibody epitope bias and onset of celiac disease. Proc. Natl Acad. Sci. USA 116, 15134–15139 (2019).
Chen, X. et al. Structural basis for antigen recognition by transglutaminase 2-specific autoantibodies in celiac disease. J. Biol. Chem. 290, 21365–21375 (2015).
Hnida, K. et al. Epitope-dependent functional effects of celiac disease autoantibodies on transglutaminase 2. J. Biol. Chem. 291, 25542–25552 (2016).
Ballew, J. T. et al. Antibody biomarker discovery through in vitro directed evolution of consensus recognition epitopes. Proc. Natl Acad. Sci. USA 110, 19330–19335 (2013).
Osman, A. A. et al. B cell epitopes of gliadin. Clin. Exp. Immunol. 121, 248–254 (2000).
Aleanzi, M., Demonte, A. M., Esper, C., Garcilazo, S. & Waggener, M. Celiac disease: antibody recognition against native and selectively deamidated gliadin peptides. Clin. Chem. 47, 2023–2028 (2001).
Dørum, S. et al. Gluten-specific antibodies of celiac disease gut plasma cells recognize long proteolytic fragments that typically harbor T-cell epitopes. Sci. Rep. 6, 25565 (2016).
Lefranc, M.-P. et al. IMGT®, the international ImMunoGeneTics information system® 25 years on. Nucleic Acids Res. 43, D413–D422 (2014).
Risnes, L. F. et al. Circulating CD103+ γδ and CD8+ T cells are clonally shared with tissue-resident intraepithelial lymphocytes in celiac disease. Mucosal Immunol. 14, 842–851 (2021).
Bartolomé-Casado, R. et al. CD4+ T cells persist for years in the human small intestine and display a TH1 cytokine profile. Mucosal Immunol. 14, 402–410 (2021).
Bartolomé-Casado, R. et al. Resident memory CD8 T cells persist for years in human small intestine. J. Exp. Med. 216, 2412–2426 (2019).
Steenholt, J. V. et al. The composition of T cell subtypes in duodenal biopsies are altered in coeliac disease patients. PLoS ONE 12, e0170270 (2017).
Cianci, R. et al. Tissue-infiltrating lymphocytes analysis reveals large modifications of the duodenal “immunological niche” in coeliac disease after gluten-free diet. Clin. Transl. Gastroenterol. 3, e28–e28 (2012).
Di Sabatino, A. et al. Phenotyping of peripheral blood lymphocytes in adult coeliac disease. Immunology 95, 572–576 (1998).
Halstensen, T. S. & Brandtzaeg, P. Activated T lymphocytes in the celiac lesion: Non-proliferative activation (CD25) of CD4+ α/β cells in the lamina propria but proliferation (Ki-67) of α/β and γ/δ cells in the epithelium. Eur. J. Immunol. 23, 505–510 (1993).
Cook, L. et al. Circulating gluten-specific FOXP3+CD39+ regulatory T cells have impaired suppressive function in patients with celiac disease. J. Allergy Clin. Immunol. 140, 1592–1603.e1598 (2017).
Fernández, S. et al. Characterization of gliadin-specific Th17 cells from the mucosa of celiac disease patients. Am. J. Gastroenterol. 106, 528–538 (2011).
Christophersen, A. et al. Healthy HLA-DQ2.5+ subjects lack regulatory and memory T cells specific for immunodominant gluten epitopes of celiac disease. J. Immunol. 196, 2819–2826 (2016).
Bauer, S. et al. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science 285, 727–729 (1999).
Billadeau, D. D., Upshaw, J. L., Schoon, R. A., Dick, C. J. & Leibson, P. J. NKG2D-DAP10 triggers human NK cell–mediated killing via a Syk-independent regulatory pathway. Nat. Immunol. 4, 557–564 (2003).
Meresse, B. et al. Coordinated induction by IL15 of a TCR-independent NKG2D signaling pathway converts CTL into lymphokine-activated killer cells in celiac disease. Immunity 21, 357–366 (2004).
Mayassi, T. et al. Chronic inflammation permanently reshapes tissue-resident immunity in celiac disease. Cell 176, 967–981.e919 (2019).
Bhagat, G. et al. Small intestinal CD8+TCRgammadelta+NKG2A+ intraepithelial lymphocytes have attributes of regulatory cells in patients with celiac disease. J. Clin. Invest. 118, 281–293 (2008).
Calleja, S. et al. Dynamics of non-conventional intraepithelial lymphocytes-NK, NKT, and γδ T-in celiac disease: relationship with age, diet, and histopathology. Dig. Dis. Sci. 56, 2042–2049 (2011).
López-Palacios, N. et al. Evaluation of T cells in blood after a short gluten challenge for coeliac disease diagnosis. Dig. Liver Dis. 50, 1183–1188 (2018).
Núñez, C. & Fernández-Bañares, F. Some considerations about γδ and CD8+ T-cell responses in blood after gluten challenge in treated celiac disease. Mucosal Immunol. 14, 1214–1215 (2021).
Risnes, L. F., Eggesbø, L. M., Christophersen, A. & Sollid, L. M. Response to: “Some considerations about γδ and CD8+ T-cell responses in blood after gluten challenge in treated celiac disease”. Mucosal Immunol. 14, 1216–1217 (2021).
Pinto, D. et al. A functional BCR in human IgA and IgM plasma cells. Blood 121, 4110–4114 (2013).
Høydahl, L. S. et al. Plasma cells are the most abundant gluten peptide MHC-expressing cells in inflamed intestinal tissues from patients with celiac disease. Gastroenterology 156, 1428–1439.e1410 (2019).
Landsverk, O. J. et al. Antibody-secreting plasma cells persist for decades in human intestine. J. Exp. Med. 214, 309–317 (2017).
Freeman, H. J. Dietary compliance in celiac disease. World J. Gastroenterol. 23, 2635–2639 (2017).
Fillatreau, S. Regulatory functions of B cells and regulatory plasma cells. Biomed. J. 42, 233–242 (2019).
Santaguida, M. G. et al. Breg cells in celiac disease isolated or associated to Hashimoto’s thyroiditis. Int. J. Endocrinol. 2018, 5290865 (2018).
van Unen, V. et al. Mass cytometry of the human mucosal immune system identifies tissue- and disease-associated immune subsets. Immunity 44, 1227–1239 (2016).
Christophersen, A. et al. Distinct phenotype of CD4+ T cells driving celiac disease identified in multiple autoimmune conditions. Nat. Med. 25, 734–737 (2019).
Nair, N. et al. High-dimensional immune profiling of total and rotavirus VP6-specific intestinal and circulating B cells by mass cytometry. Mucosal Immunol. 9, 68–82 (2016).
Snir, O., Kanduri, C., Lundin, K. E. A., Sandve, G. K. & Sollid, L. M. Transcriptional profiling of human intestinal plasma cells reveals effector functions beyond antibody production. U. Eur. Gastroenterol. J. 7, 1399–1407 (2019).
Bragde, H., Jansson, U., Fredrikson, M., Grodzinsky, E. & Söderman, J. Celiac disease biomarkers identified by transcriptome analysis of small intestinal biopsies. Cell Mol. Life Sci. 75, 4385–4401 (2018).
Quinn, E. M. et al. Transcriptome analysis of CD4+ T cells in coeliac disease reveals imprint of BACH2 and IFNγ regulation. PLoS One 10, e0140049 (2015).
Wolf, J. et al. Deciphering the transcriptomic heterogeneity of duodenal coeliac disease biopsies. Int. J. Mol. Sci. 22, 2551 (2021).
Leonard, M. M. et al. RNA sequencing of intestinal mucosa reveals novel pathways functionally linked to celiac disease pathogenesis. PLoS One 14, e0215132 (2019).
Atlasy, N. et al. Single cell transcriptome atlas of immune cells in human small intestine and in celiac disease. Preprint at https://www.biorxiv.org/content/10.1101/721258v1 (2019).
Guikema, J. E. et al. CD27 is heterogeneously expressed in multiple myeloma: low CD27 expression in patients with high-risk disease. Br. J. Haematol. 121, 36–43 (2003).
Kerkman, P. et al. Generation of plasma cells and CD27-IgD- B cells during hantavirus infection is associated with distinct pathological findings. Clin. Transl. Immunol. 10, e1313 (2021).
du Pré, M. F. & Sollid, L. M. T-cell and B-cell immunity in celiac disease. Best. Pract. Res. Clin. Gastroenterol. 29, 413–423 (2015).
Crotty, S. Follicular helper CD4 T cells (TFH). Annu. Rev. Immunol. 29, 621–663 (2011).
Meresse, B., Ripoche, J., Heyman, M. & Cerf-Bensussan, N. Celiac disease: from oral tolerance to intestinal inflammation, autoimmunity and lymphomagenesis. Mucosal Immunol. 2, 8–23 (2009).
Pabst, O. & Mowat, A. M. Oral tolerance to food protein. Mucosal Immunol. 5, 232–239 (2012).
Jabri, B. & Sollid, L. M. Tissue-mediated control of immunopathology in coeliac disease. Nat. Rev. Immunol. 9, 858–870 (2009).
Kmieciak, M. et al. Human T cells express CD25 and Foxp3 upon activation and exhibit effector/memory phenotypes without any regulatory/suppressor function. J. Transl. Med. 7, 89 (2009).
Jabri, B. & Abadie, V. IL-15 functions as a danger signal to regulate tissue-resident T cells and tissue destruction. Nat. Rev. Immunol. 15, 771–783 (2015).
Meresse, B., Korneychuk, N., Malamut, G. & Cerf-Bensussan, N. Interleukin-15, a master piece in the immunological jigsaw of celiac disease. Dig. Dis. 33, 122–130 (2015).
Korneychuk, N. et al. Interleukin 15 and CD4+ T cells cooperate to promote small intestinal enteropathy in response to dietary antigen. Gastroenterology 146, 1017–1027 (2014).
Kooy-Winkelaar, Y. M. C. et al. CD4 T-cell cytokines synergize to induce proliferation of malignant and nonmalignant innate intraepithelial lymphocytes. Proc. Natl Acad. Sci. USA 114, E980–E989 (2017).
Hoytema van Konijnenburg, D. P. et al. Intestinal epithelial and intraepithelial T cell crosstalk mediates a dynamic response to infection. Cell 171, 783–794.e713 (2017).
Iversen, R., Mysling, S., Hnida, K., Jørgensen, T. J. & Sollid, L. M. Activity-regulating structural changes and autoantibody epitopes in transglutaminase 2 assessed by hydrogen/deuterium exchange. Proc. Natl Acad. Sci. USA 111, 17146–17151 (2014).
Lindfors, K., Kaukinen, K. & Maki, M. A role for anti-transglutaminase 2 autoantibodies in the pathogenesis of coeliac disease? Amino Acids 36, 685–691 (2009).
Halstensen, T. S., Hvatum, M., Scott, H., Fausa, O. & Brandtzaeg, P. Association of subepithelial deposition of activated complement and immunoglobulin G and M response to gluten in celiac disease. Gastroenterology 102, 751–759 (1992).
Sugai, E. et al. Accuracy of testing for antibodies to synthetic gliadin–related peptides in celiac disease. Clin. Gastroenterol. Hepatol. 4, 1112–1117 (2006).
Sulkanen, S. et al. Tissue transglutaminase autoantibody enzyme-linked immunosorbent assay in detecting celiac disease. Gastroenterology 115, 1322–1328 (1998).
Jenks, S. A., Cashman, K. S., Woodruff, M. C., Lee, F. E.-H. & Sanz, I. Extrafollicular responses in humans and SLE. Immunol. Rev. 288, 136–148 (2019).
Mesin, L., Sollid, L. M. & Di Niro, R. The intestinal B-cell response in celiac disease. Front. Immunol. 3, 313 (2012).
Cruikshank, W. W. et al. Molecular and functional analysis of a lymphocyte chemoattractant factor: association of biologic function with CD4 expression. Proc. Natl Acad. Sci. USA 91, 5109–5113 (1994).
Bamford, R. N., DeFilippis, A. P., Azimi, N., Kurys, G. & Waldmann, T. A. The 5′ untranslated region, signal peptide, and the coding sequence of the carboxyl terminus of IL-15 participate in its multifaceted translational control. J. Immunol. 160, 4418–4426 (1998).
Mishra, A., Sullivan, L. & Caligiuri, M. A. Molecular pathways: interleukin-15 signaling in health and in cancer. Clin. Cancer Res. 20, 2044–2050 (2014).
Nishimura, H. et al. Differential roles of interleukin 15 mRNA isoforms generated by alternative splicing in immune responses in vivo. J. Exp. Med. 191, 157–170 (2000).
Tagaya, Y. et al. Generation of secretable and nonsecretable interleukin 15 isoforms through alternate usage of signal peptides. Proc. Natl Acad. Sci. USA 94, 14444–14449 (1997).
Regev, A. et al. The Human Cell Atlas. Elife 6, e27041 (2017).
Dey, S. S., Kester, L., Spanjaard, B., Bienko, M. & van Oudenaarden, A. Integrated genome and transcriptome sequencing of the same cell. Nat. Biotechnol. 33, 285–289 (2015).
Han, K. Y. et al. SIDR: simultaneous isolation and parallel sequencing of genomic DNA and total RNA from single cells. Genome Res. 28, 75–87 (2018).
Rodriguez-Meira, A. et al. Unravelling intratumoral heterogeneity through high-sensitivity single-cell mutational analysis and parallel RNA sequencing. Mol. Cell. 73, 1292–1305.e1298 (2019).
Angermueller, C. et al. Parallel single-cell sequencing links transcriptional and epigenetic heterogeneity. Nat. Methods 13, 229–232 (2016).
Hu, Y. et al. Simultaneous profiling of transcriptome and DNA methylome from a single cell. Genome Biol. 17, 88 (2016).
Chen, S., Lake, B. B. & Zhang, K. High-throughput sequencing of the transcriptome and chromatin accessibility in the same cell. Nat. Biotechnol. 37, 1452–1457 (2019).
Cao, J. et al. Joint profiling of chromatin accessibility and gene expression in thousands of single cells. Science 361, 1380–1385 (2018).
Zhu, C. et al. An ultra high-throughput method for single-cell joint analysis of open chromatin and transcriptome. Nat. Struct. Mol. Biol. 26, 1063–1070 (2019).
Clark, S. J. et al. scNMT-seq enables joint profiling of chromatin accessibility DNA methylation and transcription in single cells. Nat. Commun. 9, 781 (2018).
Hou, Y. et al. Single-cell triple omics sequencing reveals genetic, epigenetic, and transcriptomic heterogeneity in hepatocellular carcinomas. Cell Res. 26, 304–319 (2016).
Stoeckius, M. et al. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods 14, 865–868 (2017).
Frei, A. P. et al. Highly multiplexed simultaneous detection of RNAs and proteins in single cells. Nat. Methods 13, 269–275 (2016).
Genshaft, A. S. et al. Multiplexed, targeted profiling of single-cell proteomes and transcriptomes in a single reaction. Genome Biol. 17, 188 (2016).
Peterson, V. M. et al. Multiplexed quantification of proteins and transcripts in single cells. Nat. Biotechnol. 35, 936–939 (2017).
Gerlach, J. P. et al. Combined quantification of intracellular (phospho-)proteins and transcriptomics from fixed single cells. Sci. Rep. 9, 1469 (2019).
Katzenelenbogen, Y. et al. Coupled scRNA-seq and intracellular protein activity reveal an immunosuppressive role of TREM2 in cancer. Cell 182, 872–885.e819 (2020).
Swanson, E. et al. Simultaneous trimodal single-cell measurement of transcripts, epitopes, and chromatin accessibility using TEA-seq. Elife 10, e63632 (2021).
Mimitou, E. P. et al. Scalable, multimodal profiling of chromatin accessibility, gene expression and protein levels in single cells. Nat. Biotechnol. (2021).
Mimitou, E. P. et al. Multiplexed detection of proteins, transcriptomes, clonotypes and CRISPR perturbations in single cells. Nat. Methods 16, 409–412 (2019).
Dahal-Koirala, S., Risnes, L. F. & Sollid, L. M. Pathogenesis of coeliac disease - a disorder driven by gluten-specific CD4+ T cells. In Coeliac Disease and Gluten-related Disorders (eds. Schieptti, A. & Sanders, D.) (Academic Press, London, 2021 [in press]).
Acknowledgements
This was supported by grants from Stiftelsen KG Jebsen (project SKGJ-MED-017), the South-Eastern Norway Regional Health Authority (project 2020027) and the University of Oslo World-leading research program on human immunology (WL-IMMUNOLOGY). We thank Shiva Dahal-Koirala for critically commenting on the manuscript.
Author information
Authors and Affiliations
Contributions
I.L. and L.M.S. drafted and revised the manuscript. I.L. created the figures.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Lindeman, I., Sollid, L.M. Single-cell approaches to dissect adaptive immune responses involved in autoimmunity: the case of celiac disease. Mucosal Immunol 15, 51–63 (2022). https://doi.org/10.1038/s41385-021-00452-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41385-021-00452-0