Abstract
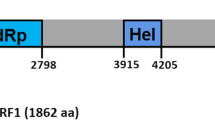
The complete genome sequence of a double-stranded RNA (dsRNA) virus, Rhizoctonia solani dsRNA virus 11 (RsRV11), isolated from Rhizoctonia solani AG-1 IA strain 9-11 was determined. The RsRV11 genome is 9,555 bp in length and contains three conserved domains: structural maintenance of chromosomes (SMC) superfamily, phosphoribulokinase (PRK), and RNA-dependent RNA polymerase (RdRp). The RsRV11 genome has two non-overlapping open reading frames (ORFs). ORF1 is predicted to encode a 204.12-kDa protein that shares low but significant amino acid sequence similarity with a putative protein encoded by Rhizoctonia solani RNA virus HN008 (RsRV-HN008). ORF2 potentially encodes a 132.41-kDa protein that contains the conserved domain of the RdRp. Phylogenetic analysis indicated that RsRV11 clustered with RsRV-HN008 in a separate clade from other virus families. This implies that RsRV11 and RsRV-HN008 should be included in a new mycovirus taxon close to the family Megabirnaviridae and that RsRV11 is a new mycovirus.


Similar content being viewed by others
References
Ghabrial SA, Castón JR, Jiang D, Nibert ML, Suzuki N (2015) 50-plus years of fungal viruses. Virology 479:356–368
Xie J, Jiang D (2014) New insights into mycoviruses and exploration for the biological control of crop fungal diseases. Annu Rev Phytopathol 52:45–68
Son M, Yu J, Kim KH (2015) Five questions about mycoviruses. PLoS pathog 11:e1005172
Nuss DL (2005) Hypovirulence: mycoviruses at the fungal–plant interface. Nat Rev Microbiol 3:632–642
Griffith JL, Coleman LE, Raymond AS, Goodson SG, Pittard WS, Tsui C, Devine SE (2003) Functional genomics reveals relationships between the retrovirus-like Ty1 element and its host Saccharomyces cerevisiae. Genetics 164:867–879
Ogoshi A (1996) Introduction – the genus Rhizoctonia. In. Rhizoctonia species: taxonomy, molecular biology, ecology, pathology and disease control. Eds. Sneh B, Jabaji-Hare S, Neate S and Dijst G. Kluwer Academic Publishers, The Netherlands, pp 1-9
Kuhn J (1858) Die Krankenheiten der Kulturwachse, ihre Ursachen und ihre Verhutung. Gustav Bosselman, Berlin, p 312
Anderson NA (1982) The genetics and pathology of Rhizoctonia solani. Annu Rev Phytopathol 20:329–347
Zheng L, Liu H, Zhang M, Cao X, Zhou E (2013) The complete genomic sequence of a novel mycovirus from Rhizoctonia solani AG-1 IA strain B275. Arch Virol 158:1609–1612
Zheng L, Zhang M, Chen Q, Zhu M, Zhou E (2014) A novel mycovirus closely related to viruses in the genus Alphapartitivirus confers hypovirulence in the phytopathogenic fungus Rhizoctonia solani. Virology 456–457:220–226
Zhang M, Zheng L, Liu C, Shu C, Zhou E (2018) Characterization of a novel dsRNA mycovirus isolated from strain A105 of Rhizoctonia solani AG-1 IA. Arch Virol 163:427–430
Liu C, Zeng M, Zhang M, Shu C, Zhou E (2018) Complete nucleotide sequence of a Partitivirus from Rhizoctonia solani AG-1 IA Strain C24. Viruses 10:703
Zheng L, Shu C, Zhang M, Yang M, Zhou E (2019) Molecular characterization of a novel Endornavirus conferring hypovirulence in rice sheath blight fungus Rhizoctonia solani AG-1 IA strain GD-2. Viruses 11:178
Darissa O, Willingmann P, Adam G (2010) Optimized approaches for the sequence determination of double-stranded RNA templates. J Virol Methods 169:397–403
Morris TJ, Dodds JA (1979) Isolation and analysis of double stranded RNA from virus-infected plant and fungal tissue. Phytopathology 69:854–858
Froussard P (1992) A random-PCR method (rPCR) to construct whole cDNA library from low amounts of RNA. Nucleic Acids Res 20:2900
Potgieter AC, Page NA, Liebenberg J, Wright IM, Landt O, van Dijk AA (2009) Improved strategies for sequence-independent amplification and sequencing of viral double-stranded RNA genomes. J Gen Virol 90:1423–1432
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The CLUSTAL X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Soppa J, Kobayashi K, Noirot-Gros MF, Oesterhelt D, Ehrlich SD, Dervyn E, Moriya S (2002) Discovery of two novel families of proteins that are proposed to interact with prokaryotic SMC proteins, and characterization of the Bacillus subtilis family members ScpA and ScpB. Mol Microbiol 45:59–71
Hirano M, Hirano T (2002) Hinge-mediated dimerization of SMC protein is essential for its dynamic interaction with DNA. EMBO J 21:5733–5744
Jensen RB, Shapiro L (2003) Cell-cycle-regulated expression and subcellular localization of the Caulobacter crescentus SMC chromosome structural protein. J Bacteriol 185:3068–3075
Liu L, Xie J, Cheng J, Fu Y, Li G, Yi X, Jiang D (2014) Fungal negative-stranded RNA virus that is related to bornaviruses and nyaviruses. Proc Natl Acad Sci 111:12205–12210
Jiāng D, Ayllón MA, Marzano SYL (2019) ICTV virus taxonomy profile: Mymonaviridae. J Gen Virol 100:1343–1344
Li P, Zhang H, Chen X, Qiu D, Guo L (2015) Molecular characterization of a novel hypovirus from the plant pathogenic fungus Fusarium graminearum. Virology 481:151–160
Li P, Bhattacharjee P, Wang S, Zhang L, Ahmed I, Guo L (2019) Mycoviruses in Fusarium species: an update. Front Cell Infect Microbiol 9:257
Gilbert K, Holcomb EE, Allscheid RL, Carrington J (2019) Discovery of new mycoviral genomes within publicly available fungal transcriptomic datasets. BioRxiv. https://doi.org/10.1101/510404
Sato Y, Miyazaki N, Kanematsu S, Xie J, Ghabrial SA, Hillman BI, Suzuki N (2019) Ictv Report Consortium. ICTV Virus Taxonomy Profile: Megabirnaviridae. J Gen Virol 100:1269–1270
Petrzik K, Sarkisova T, Starý J, Koloniuk I, Hrabáková L, Kubešová O (2016) Molecular characterization of a new monopartite dsRNA mycovirus from mycorrhizal Thelephora terrestris (Ehrh.) and its detection in soil oribatid mites (Acari: Oribatida). Virology 489:12–19
Li Y, Xu P, Zhang L, Xia Z, Qin X, Yang G, Mo X (2015) Molecular characterization of a novel mycovirus from Rhizoctonia fumigata AG-Ba isolate C-314 Baishi. Arch Virol 160:2371–2374
Kozlakidis Z, Hacker CV, Bradley D, Jamal A, Phoon X, Webber J, Brasier CM, Buck KW, Coutts RH (2009) Molecular characterisation of two novel double-stranded RNA elements from Phlebiopsis gigantea. Virus Genes 39:132–136
Ohta C, Taguchi T, Takahashi S, Ohtsuka K, Eda K, Ayusawa S, Magae Y (2008) Detection of double stranded RNA elements in cultivated Lentinula edodes (in Japanese). Mushroom Sci Biotechnol. 16:155–158
Magae Y (2012) Molecular characterization of a novel mycovirus in the cultivated mushroom, Lentinula edodes. Virol. J. 9:1–6
Acknowledgements
This research was supported the National Natural Scientific Foundation of China (31960525, 31160352, 31360423) and grants-in-aid from the Scientific Research Project of Kunming University (YJL16006) and the General Program of University Union in Yunnan Province (2018FH 001-032).
Author information
Authors and Affiliations
Corresponding author
Additional information
Handling Editor: Massimo Turina.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sun, Y., Li, Y., Dong, W. et al. Molecular characterization of a novel mycovirus isolated from Rhizoctonia solani AG-1 IA strain 9-11. Arch Virol 166, 3229–3232 (2021). https://doi.org/10.1007/s00705-021-05219-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-021-05219-3