Abstract
Our study attempted to explore the mechanism underlying the role of LuxR family transcriptional regulator abaR in biofilm formation by Acinetobacter baumannii. The abaR gene was knocked out in ATCC 17978 strain using homologous recombination method. The growth curve and biofilm formation were measured in the wild type and abaR gene knockdown strains. Transcriptome sequencing was performed in the wild type and abaR gene knockdown strains following 8 h of culture. The growth curve in the abaR gene knockdown strain was similar to that of the wild-type strain. Biofilm formation significantly declined in the abaR gene knockdown strain at 8 and 48 h after culture. A total of 137 differentially expressed genes (DEGs) were obtained including 20 downregulated DEGs and 117 upregulated DEGs. Genes with differential expression were closely related to viral procapsid maturation (GO:0046797), acetoin catabolism (GO:0045150), carbon metabolism (ko01200), and the glycolysis/gluconeogenesis (ko00010)-related pathways. The results of the eight verified expression DEGs were consistent with the results predicted by bioinformatics. AbaR gene knockdown significantly affected biofilm formation by A. baumannii ATCC 17978 strain. The glycolysis/gluconeogenesis pathways were significantly dysregulated and induced by abaR gene knockdown in A. baumannii.
Similar content being viewed by others
Introduction
Acinetobacter baumannii is a Gram-negative coccobacillus which is troublesome pathogen in hospitalized patients globally. A. baumannii has emerged as a major challenge in infected patients because of its multiple drug resistance [1, pp. 939–951]. Recent data show that A. baumannii is one of the top four most common pathogens isolated in the hospital environment between 2005 and 2017 in China [2, pp. S128–S134]. In global intensive care units (ICUs), the infection rate of A. baumannii accounts for 20% of the total number of infections [3, pp. 25–36]. The resistance profile of Acinetobacter spp. indicates that the resistance rate against the five commonly used antimicrobials, except amikacin, is increasing. The resistance rate of A. baumannii against carbapenems varied from 18 to 25% in different provinces of China [2, pp. S128–S134]. A. baumannii can cause a variety of nosocomial infections, most of which involve the respiratory tract and some involve skin wound infections and bacteremia [4]. This is corroborated by the report that A. baumannii is an increasing threat in burn centers because of its high multidrug resistance based on clinical data obtained from a comprehensive hospital covering 2007–2013 and the CHINET (the surveillance system for bacterial epidemiology and resistance in China) data [5, pp. 1718–1719].
Considerable attention has been focused on exploring the mechanism of drug resistance in A. baumannii in clinical settings [6]. One of the major factors associated with bacterial resistance to antimicrobials is the ability to form biofilms. Previous studies have shown that strains that can form biofilms are more resistant to antibiotics than strains that cannot [7]. Recent evidence shows that biofilm formation is closely related to drug resistance and virulence in A. baumannii [8, pp. 119–127]. It is reported that the biofilm-forming ability of A. baumannii in a solid–liquid interface is approximately three times higher than that of other Acinetobacter species [9, pp. 1–4]. A. baumannii can rapidly produce biofilms while forming antibiotic resistance, and the level of biofilm-specific resistance can vary according to the induction response of the biofilm population and the action mechanism of antibiotics [10, p. 817]. Cold plasma technology, as a new tool for purification of biofilm-contaminated surfaces, has received more and more clinical attention. The increase in the amount of A. baumannii biofilm could reduce the antibacterial effect of cold plasma, thereby improving its tolerance to cold plasma exposure [11, pp. 344–349]. The biofilm formation characteristics of A. baumannii are related to the expression of genes. The genes that have been reported to be associated with the formation of A. baumannii biofilm include csu locus, bap, abaI, adeFGH, and ompA [7]. csuE encodes a fimbriae assembly system, and the formation of fimbriae is an important step in the formation of biofilm on non-living surfaces [12, pp. 3473–3484]. Bap can support the development of mature biofilm structures [13, pp. 1036–1044]. The mutation of abaI will cause the failure of the acyl-homoserine lactone signals to be generated and further damage the maturation of the biofilm [14, pp. 951–957]. adeFGH is involved in the synthesis and transport of autoinducer molecules during the formation of biofilms [15, pp. 4817–25]. ompA encodes a porin protein involved in the formation of biofilms [16, pp. 3150–3160]. Previous reports have implicated the abaR gene in A. baumannii biofilm formation and disruption of abaR expression is associated with a decline in biofilm formation [17, pp. 1802–1805; 18, pp. 200–205; 19, pp. 1339–1346]. Although there have been advances in exploring the mechanism underlying biofilm formation, the role of abaR at the transcriptome level has not been clarified completely.
Therefore, in this study, we aimed to determine the role of abaR in A. baumannii biofilm formation through abaR knockdown and explore the mechanism regulated by abaR expression using high-throughput transcriptional sequencing.
Materials and Methods
Bacterial Strains and abaR Gene Knockouts
A. baumannii ATCC 17978 strain was obtained from Shanghai Santa Biotechnology Co. Ltd., Shanghai, China. The ATCC 17978 strain was cultured in Luria–Bertani (LB) medium at 37 °C overnight. The abaR gene knockdown was performed using homologous recombination method [20, pp. 3743–3751]. A recombinant DNA fragment (ΔabaR::Kn) consisting of kanamycin resistance gene (Kn) sequences (obtained from pKD4 plasmid using PCR) and the upper and lower homologous recombination arms for abaR gene (amplified from ATCC17978 genome using PCR) were obtained using splicing PCR technology. ΔabaR::Kn target fragments were amplified by transferring them into pUC19 vector and tested by PCR analysis. The primers used are listed in Supplementary Table 1. Next, ΔabaR::Kn sequences were linked to pCVD442 vector and transferred to Escherichia coli β2155 by electrotransformation. Conjugation between ATCC17978 strain and the E. coli was performed and the knockdown ATCC17978 strains were selected for kanamycin resistance [21, pp. 71–76].
Construction of Complementary Strains
To complement the knockout mutant, the target gene abaR was amplified from the cDNA of ATCC17978 strain, and the amplification product was cloned into the vector pBAD33-TCR to construct a pBAD33-TCR-abaR. The vector pBAD33-TCR has the origin of replication ori amplified on the pWH1266 plasmid and the tetracycline resistance gene; so, this plasmid can be amplified in A. baumannii. Then, pBAD33-TCR-abaR was transformed to E. coli competent cells DH5α. The plasmids that were positive after tetracycline screening were sequenced. Then, the plasmids with correct sequencing results were transferred to E. coli β2163 competent cells and the positive clone E. coli β2163-[pBAD33-TCR-abaR] was screened. Finally, E. coli β2163-[pBAD33-TCR-abaR] was transformed into knockdown ATCC17978 strains using electrotransformation method [22, pp. 162–168] and the recombinant clones were screened out, followed by PCR verification, and PCR products are sequenced for sequence confirmation (Supplementary Fig. 1). The primers used in this section were listed in Supplementary Table 2.
Growth Curve Analysis
ATCC 17978 and abaR knockdown strains at 1 × 108 CFU/mL were maintained in fresh LB media at 37 °C for consecutive 18 h. The optical density (OD) of cultured strains in each group was evaluated for each hour using a Microplate reader (Bio-Rad, Hercules, CA, USA). There were three repeats in each group.
Biofilm Formation Analysis
Next, 200 μL of strain cultures (1 × 108 CFU/mL) was seeded onto each well of a 96-well plate. After culturing for 8, 24, and 48 h, the cultures were washed thrice with 200 μL PBS. Next, the cultures were fixed in methanol for 15 min followed by staining with 0.2% crystal violet solution for 20 min. After washing, 200 μL of 33% glacial acetic acid was added to each well and then, OD values were measured using a Microplate reader.
RNA Sequencing
After culturing for 8 h, ATCC17978 and abaR knockdown strains were collected for further analysis. Total RNA of the cultures in the two groups were isolated using mirVana miRNA isolation kit (Ambion) following the manufacturer’s protocols, separately [23, p. e72968]. Because the vast majority of RNA in bacteria is rRNA, rRNA was obtained using Ribo-Zero Magnetic Kit (MRZB12424, Illumina, CA, USA) and removed, following which the mRNAs were broken into fragments as templates for cDNA synthesis [24]. An RNA library was constructed using PCR analysis. The quality and size of the DNA fragments were evaluated using Agilent 2100 Bioanalyzer. The whole transcriptome was then sequenced based on the Illumina platform.
Data Processing
The raw data were downloaded and quality control was performed, using Trimmomatic software, including removing adaptor and low-quality reads. Clean reads were then mapped to reference genome using Rockhooper2 software. The gene expression level was measured based on the number of reads mapped to the genome and exome according to the reads per kilobase per million mapped reads (RPKM) method. The formula is provided in Eq. (1) as follows:
Differentially Expressed Gene Analysis
The gene counts were normalized using DESeq software to produce the significant genes with differential expression between the abaR knockdown ATCC 17978 and wild-type ATCC 17978 strains. The fold change (FC) of gene expression was calculated and negative binomial distribution test was applied to evaluate the significance of the differences in gene reads between two samples. The log|FC|> 1.5 and P value < 0.05 were set as the cutoff values.
Function Enrichment Analysis
The Gene Ontology (GO) project provides the possibility of biological function annotation of genes of interest [25, pp. D322–D326]. The differentially expressed genes were subjected to GO function analysis and the significant P values were tested using the hypergeometric distribution test. GO terms with P values < 0.05 were considered significant.
Pathway Enrichment Analysis
The Kyoto Encyclopedia of Genes and Genomes (KEGG) (http://www.genome.ad.jp/kegg/) is a pathway-related database [26, pp. 91–100]. KEGG pathway enrichment analysis was performed to explore the pathways involved in differentially expressed genes using the DAVID online tool. The enrichment P values were calculated using the hypergeometric distribution test. A P value < 0.05 was considered the cutoff value.
Real-Time Reverse Transcription-PCR Verification
RT-PCR was carried out to determine the relative expression of some DEGs. Total RNA was isolated from wild-type strains, mutant strains, and complementary strains after 8 h of culture. The concentration of RNA was detected using NanoDrop 2000. RNA was reverse-transcribed to cDNA using TransScript All-in-One First-Strand cDNA Synthesis SuperMIX for qPCR kit. PerfectStartTM Green qPCR SuperMix kit and LightCycler® 480 system were used together to perform RT-PCR. All assays were performed in triplicate. The relative expression levels of RNAs were calculated using the 2−ΔΔCt method. 16 s was set as internal control. The primer sequences are listed in Supplementary Table 3.
Statistical Analysis
The data for strain growth and biofilm formation are expressed as mean ± standard deviation (SD). Multiple group comparison was performed using one-way ANOVA analysis and the difference between two groups was analyzed using the LSD test. P < 0.05 was considered significant.
Deposit the RNA-Seq Data
The raw sequencing data were deposited in the National Center for Biotechnology Information (NCBI) Sequence ReadArchive (SRA) database under accession number PRJNA720897.
Results
Effective abaR Gene Knockouts in ATCC 17978 Strain
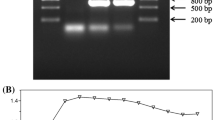
PCR analysis of a recombinant DNA fragment (ΔabaR::Kn) showed that the size of the target sequence was 3029 bp (Fig. 1A). Next, the knockdown ATCC17978 strain was selected for kanamycin resistance. The PCR amplification of the target fragment was about 3300 bp, which indicated that the abaR gene sequence was successfully replaced by ΔabaR::Kn target fragment (Fig. 1B).
Growth Curve of ATCC 17978 Strain and abaR Knockdown Strain
The strain growth curve in Fig. 2 showed that the OD values for the ATCC 17978 strain were slightly higher than the abaR knockdown strain at 2, 3, and 4 h of culture. No significant differences were observed in the OD values of the two strain groups from 5 to 12 h of culture. All strains entered the plateau stage after 14 h of culture. ATCC17978 entered the exponential growth stage during 3–4 h of culture in vitro and entered the plateau stage at 14 h, which was similar to the growth curve of the abaR knockdown strain.
Biofilm Formation Analysis
As shown in Fig. 3, biofilm formation by the ATCC 17978 strain increased in a time-dependent manner and peaked at 48 h of culture. However, there were no significant changes in the biofilm-forming ability of abaR knockdown strains during 0–8 h of culture. The biofilm-forming ability in the wild type and abaR knockdown strains was similar at 0–4 h of culture. At 8, 24, and 48 h after strain culture, the OD value of ATCC 17978 was 0.63 ± 0.06, 0.522 ± 0.18, and 1.04 ± 0.32, respectively. After abaR gene knockdown, the OD value was 0.41 ± 0.20, 0.45 ± 0.11, and 0.57 ± 0.02 at 8, 24, and 48 h, respectively. There were significant differences in OD values at 8 and 48 h of culture between the groups (all P values < 0.05), indicating that biofilm formation significantly declined in the ATCC 17978 strain after abaR knockdown.
Identification of Differentially Expressed Genes
With the cutoff value of log|FC|> 1.5 and P value < 0.05, a total of 137 differentially expressed genes (DEGs) were obtained between the wild type and abaR knockdown strains, including 20 downregulated genes and 117 upregulated genes (Supplementary Table 4). The volcano plot of genes with differential expression is shown in Fig. 4.
Volcano plot for differentially expressed genes in abaR knockdown strain, compared with wide-type strain. Red spots represent genes that were significantly upregulated. Green spots represent genes that were significantly downregulated. Gray spots represent genes that were not significantly different between the two groups (Color figure online)
Significant Biological Processes and Pathways for Differentially Expressed Genes
GO analysis was performed for all the genes with differential expression. A total of 79 GO terms were enriched by differentially expressed genes, among which 58 GO terms were significantly enriched with P < 0.05. The differentially expressed genes were closely related to acetoin catabolic process (GO:0045150, BP), glycolytic process (GO:0006096, BP), tartrate dehydrogenase activity (GO:0009027, MF), and viral capsid (GO:0019028, CC). A total of 22 pathways were significantly enriched and the top 15 significant pathways are listed in Table 1. The results showed that pathways such as glycolysis/gluconeogenesis (ko00010), pyruvate metabolism (ko00620), and glucagon signaling pathway (ko04922) were dysregulated in abaR knockdown strains compared with the wild-type ATCC 17978.
RT-PCR Detection
The relative expression levels of four significantly up-regulated DEGs and four significantly down-regulated DEGs were detected by PT-PCR between wild-type ATCC17978 strains and abaR knockdown strains (Fig. 5). As we predicted, compared in the wild-type ATCC17978 strains, the relative expression of A1S_3544, A1S_0849, A1S_3867, and A1S_3011 showed a significantly higher levels in the abaR knockdown strains (P < 0.001). The expression levels of A1S_3661, A1S_1709, A1S_2237, and A1S_2457 were significantly higher in the wild-type ATCC17978 strains than that in the abaR knockdown strains (P < 0.001).
We also detected the relative expression of A1S_0849 and A1S_2237 between wild-type ATCC17978 strains, abaR knockdown strains, and complementary strains (Fig. 6). The results revealed that there were no significant difference between ATCC17978 strains and complementary strains in the A1S_0849 and A1S_2237 expression. This indicates that our complementation experiment successfully restored gene expression to the wild-type level.
Discussion
A. baumannii has emerged as an important nosocomial pathogen mainly affecting patients with impaired host defences [27, pp. 640–645]. Now, A. baumannii is one of the most prevalent pathogens in burn-induced infections because of its multidrug-resistance, which can even induce bloodstream infections. A. baumannii is responsible for a vast array of infections, of which ventilator-associated pneumonia and bloodstream infections are the most common, and mortality rates can reach 35% [28, pp. 292–301]. Multidrug resistance may complicate the treatment of serious infections. Biofilm-forming ability has been found to be the major factor affecting drug resistance by A. baumannii [29, pp. 626–633]. Many gram-negative pathogens control their virulence through a quorum sensing (QS) system [30, p. 163]. AbaR is one of the QS-related genes, which is reported to play a regulatory role in biofilm formation [31, pp. 15–27]. In this study, we aimed to explore the mechanism underlying the role of abaR in A. baumannii biofilm formation.
We first determined the regulatory role of abaR in A. baumannii biofilm formation by gene knockout. In A. baumannii, abaR- and abaI-mediated QS plays a regulatory role in biofilm formation and surface motility [32, pp. 1719–1728]. The abaR gene at a size of 717 bp is localized at 1255 bp of the downstream of abaI gene. Additionally, we constructed an abaR gene knockout model in neurons using homologous recombination method. After abaR gene knockdown, the alternative target fragment in ATCC 17978 was determined to be 3300 bp using PCR analysis, which was similar to the size of ΔabaR::Kn target fragment, suggesting that the abaR gene was successfully knocked out.
Data of the growth curve analysis showed that after 4 h of culture, the growth of the wild-type ATCC 17978 entered the exponential growth phase and entered the plateau phase at 14 h after culture. The abaR knockdown strain showed a similar growth curve, which indicates that no significant effect was observed in the growth of ATCC 17978 strain following abaR knockdown.
A previous study suggested that the absorbance value of drug-resistant A. baumannii strain was similar to sensitive strains at all culture time points. Compared with the controls, the biofilm-forming ability of the ATCC 17978 strain exposed to N-Heptanoyl-l-Homoserine lactone significantly declined at 12 h of culture and that of strains exposed to N-(3-Hydroxydodecanoyl)-dl-Homoserine lactone noticeably increased [18, pp. 200–205]. Therefore, inhibition or activation of the abaR gene can influence biofilm formation. Our data showed that biofilm-forming ability significantly increased in the wild-type ATCC 17978 strain compared with the abaR knockdown ATCC 17978 strain at 8 h of culture, which was consistent with the previous findings. All these results suggest that abaR knockdown affected the biofilm-forming ability of A. baumannii without influencing strain growth.
In addition, the mechanism of the declined biofilm-forming ability of A. baumannii induced by abaR knockdown has not been clarified completely. Thus, we performed transcriptome sequencing to identify the changed gene expression profile caused by abaR knockdown. Based on the results, a total of 137 genes were differentially expressed between the wild type and abaR knockdown strains. GO analysis showed that viral procapsid maturation was closely related to the dysregulated expressed genes induced by abaR knockdown. It is reported that viral procapsid maturation is a dynamic process for viral infection [33, pp. 423–439]. The virus capsid assembly and DNA packaging have been proposed to be the target for antiviral therapy [34]. Thus, biofilm formation may share a similar group of involved genes with viral procapsid maturation and biofilm formation may be suggested as the target for antimicrobial treatment.
A previous study suggested that the changes in gene expression were more obvious in the early stage of biofilm formation than the later stages. In our study, biofilm formation was significantly changed at 8 and 24 h of culture in abaR knockdown strains compared with the wild-type strains. Thus, we performed transcriptome sequencing for the strains cultured for 8 h. Previous evidence showed that carbohydrate and amino acid metabolism were the necessary pathways in biofilm formation [35, 36, p. 13160]. Our study suggested that the DEGs in abaR knockdown strains, compared with the wild-type ATCC 17978, were significantly enriched in carbon metabolism and valine, leucine, and isoleucine degradation, which is consistent with previous reports and indicates that our findings are significant. Additionally, glycolysis and gluconeogenesis were the most significant pathways involved in the differentially expressed genes. A recent study reported that genes that play a regulatory role in gluconeogenesis are significantly upregulated in biofilm cells compared with free cells [37, p. 139]. It is reported that carbohydrates are the major composition of biofilms [38]. Carbon sources play a necessary role in the survival, growth, and infection of strains [39, pp. 2034–2045]. Accumulating evidence shows that Candida albicans can utilize glycolysis and gluconeogenesis alternatively for energy production [40]. Glycolysis plays a necessary role in the morphogenesis and virulence of Candida species. The evidence for the significant role of glycolysis and gluconeogenesis pathways in A. Baumannii virulence has been rarely reported. Thus, we suspect that the dysregulation of the glycolysis and gluconeogenesis pathways was induced by abaR knockdown in A. Baumannii.
Conclusion
Although this study lacks certain mechanism experiments, our verification of DEGs through RT-PCR showed that our bioinformatics analysis has a certain degree of credibility. Of course, further research on the protein level of these DEGs is also necessary in the future. In summary, abaR knockdown significantly decreased the biofilm-forming ability of the A. baumannii ATCC 17978 strain without affecting its growth. Carbon metabolism-related pathways, especially the glycolysis and gluconeogenesis pathways, were significantly dysregulated by abaR knockdown in A. Baumannii.
Data Availability
The data used to support the findings of this study are available from the corresponding author upon request.
Code Availability
Not applicable.
References
Dijkshoorn L, Nemec A, Seifert H (2007) An increasing threat in hospitals: multidrug-resistant Acinetobacter baumannii. Nat Rev Microbiol 5:939–951
Hu F, Zhu D, Wang F et al (2018) Current status and trends of antibacterial resistance in China. Clin Infect Dis 67:S128–S134
Joshi SG, Litake GM (2013) Acinetobacter baumannii: an emerging pathogenic threat to public health. World J Clin Infect Dis 3:25–36
Vázquez-López R, Solano-Gálvez SG, Juárez Vignon-Whaley JJ (2020) Acinetobacter baumannii resistance: a real challenge for clinicians. Antibiotics (Basel) 9:205
Guo H, Qin J, Xiang J (2016) Surveillance for and susceptibility of Acinetobacter baumannii in a large hospital and burn center in Shanghai, China, 2007–2013. Am J Infect Control 44:1718–1719
Trebosc V, Gartenmann S, Tötzl M et al (2019) Dissecting colistin resistance mechanisms in extensively drug-resistant Acinetobacter baumannii clinical isolates. mBio. https://doi.org/10.1128/mBio.01083-19
Yang CH, Su PW, Moi SH et al (2019) Biofilm formation in acinetobacter baumannii: genotype-phenotype correlation. Molecules (Basel, Switzerland) 24:1849
Longo F, Vuotto C, Donelli G (2014) Biofilm formation in Acinetobacter baumannii. New Microbiol 37:119–127
Martí S, Rodríguez-Baño J, Catel-Ferreira M et al (2011) Biofilm formation at the solid-liquid and air-liquid interfaces by Acinetobacter species. BMC Res Notes 4:1–4
Shenkutie AM, Yao MZ, Siu GK-h et al (2020) Biofilm-induced antibiotic resistance in clinical Acinetobacter baumannii isolates. Antibiotics 9:817
Flynn PB, Graham WG, Gilmore BF (2019) Acinetobacter baumannii biofilm biomass mediates tolerance to cold plasma. Lett Appl Microbiol 68:344–349
Tomaras AP, Dorsey CW, Edelmann RE et al (2003) Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: involvement of a novel chaperone-usher pili assembly system. Microbiology (Reading, England) 149:3473–3484
Loehfelm TW, Luke NR, Campagnari AA (2008) Identification and characterization of an Acinetobacter baumannii biofilm-associated protein. J Bacteriol 190:1036–1044
Piperaki ET, Tzouvelekis LS, Miriagou V et al (2019) Carbapenem-resistant Acinetobacter baumannii: in pursuit of an effective treatment. Clin Microbiol Infect 25:951–957
He X, Lu F, Yuan F et al (2015) Biofilm formation caused by clinical Acinetobacter baumannii isolates is associated with overexpression of the AdeFGH efflux pump. Antimicrob Agents Chemother 59:4817–4825
Gaddy JA, Tomaras AP, Actis LA (2009) The Acinetobacter baumannii 19606 OmpA protein plays a role in biofilm formation on abiotic surfaces and in the interaction of this pathogen with eukaryotic cells. Infect Immun 77:3150–3160
Chow JY, Yang Y, Tay SB et al (2014) Disruption of biofilm formation by the human pathogen Acinetobacter baumannii using engineered quorum-quenching lactonases. Antimicrob Agents Chemother 58:1802–1805
Guo HN, Xiang J (2017) Influences of abaR gene on biofilm formation of Acinetobacter baumannii. Zhonghua Shao Shang Za Zhi 33:200–205. https://doi.org/10.3760/cma.j.issn.1009-2587.2017.04.003
Oh MH, Han K (2020) AbaR is a LuxR type regulator essential for motility and the formation of biofilm and pellicle in Acinetobacter baumannii. Genes Genomics 42:1339–1346
Arroyo LA, Herrera CM, Fernandez L et al (2011) The pmrCAB operon mediates polymyxin resistance in Acinetobacter baumannii ATCC 17978 and clinical isolates through phosphoethanolamine modification of lipid A. Antimicrob Agents Chemother 55:3743–3751
Chhetri G, Pandey T, Chinta R et al (2015) An improved method for high-level soluble expression and purification of recombinant amyloid-beta peptide for in vitro studies. Protein Expr Purif 114:71–76. https://doi.org/10.1016/j.pep.2015.05.015
Du P, Liu D, Song H et al (2020) Novel IS26-mediated hybrid plasmid harbouring tet(X4) in Escherichia coli. J Global Antimicrobial Resistance 21:162–168
Rumbo-Feal S, Gómez MJ, Gayoso C et al (2013) Whole transcriptome analysis of Acinetobacter baumannii assessed by RNA-sequencing reveals different mRNA expression profiles in biofilm compared to planktonic cells. PLoS ONE 8:e72968
Culviner PH, Guegler CK, Laub MT (2020) A simple, cost-effective, and robust method for rRNA depletion in RNA-sequencing studies. mBio. https://doi.org/10.1128/mBio.00010-20
Consortium GO (2006) The gene ontology (GO) project in 2006. Nucleic Acids Res 34:D322–D326
Kanehisa M (2002) The KEGG database. Novartis Foundation Symposium. Wiley, Hoboken, pp 91–100
Hu Y-F, Hou CJ-Y, Kuo C-F et al (2017) Emergence of carbapenem-resistant Acinetobacter baumannii ST787 in clinical isolates from blood in a tertiary teaching hospital in Northern Taiwan. J Microbiol Immunol Infect 50:640–5
Antunes LC, Visca P, Towner KJ (2014) Acinetobacter baumannii: evolution of a global pathogen. Pathogens Dis 71:292–301
Nor A’shimi MH, Alattraqchi AG, Mohd Rani F et al (2019) Biocide susceptibilities and biofilm-forming capacities of Acinetobacter baumannii clinical isolates from Malaysia. J Infect Dev Ctries 13:626–33
Vadakkan K, Vijayanand S, Hemapriya J et al (2019) Quorum sensing inimical activity of Tribulus terrestris against gram negative bacterial pathogens by signalling interference. 3 Biotech 9:163
Saipriya K, Swathi CH, Ratnakar KS et al (2020) Quorum-sensing system in Acinetobacter baumannii: a potential target for new drug development. J Appl Microbiol 128:15–27. https://doi.org/10.1111/jam.14330
Stacy DM, Welsh MA, Rather PN et al (2012) Attenuation of quorum sensing in the pathogen Acinetobacter baumannii using non-native N-Acyl homoserine lactones. ACS Chem Biol 7:1719–1728
Cardone G, Heymann JB, Cheng N et al (2012) Procapsid assembly, maturation, nuclear exit: dynamic steps in the production of infectious herpesvirions. In: Rossmann MG, Rao VB (eds) Viral molecular machines. Springer, Boston, pp 423–439
Zhang X, Jia R, Zhou J et al (2016) Capsid-targeted viral inactivation: a novel tactic for inhibiting replication in viral infections. Viruses 8:258
Lemos JA, Palmer SR, Zeng L et al (2019) The biology of Streptococcus mutans. Microbiol Spectr 7(1):1–26. https://doi.org/10.1128/microbiolspec.GPP3-0051-2018
Lu H, Que Y, Wu X et al (2019) Metabolomics deciphered metabolic reprogramming required for biofilm formation. Sci Rep 9:13160–13166
Li Z, Chen Y, Liu D et al (2015) Involvement of glycolysis/gluconeogenesis and signaling regulatory pathways in Saccharomyces cerevisiae biofilms during fermentation. Front Microbiol 6:139
Jara J, Alarcón F, Monnappa AK et al (2021) Self-adaptation of pseudomonas fluorescens biofilms to hydrodynamic stress. Front Microbiol. https://doi.org/10.3389/fmicb.2020.588884
Carlson HK, Lui LM, Price MN et al (2020) Selective carbon sources influence the end products of microbial nitrate respiration. ISME J 14:2034–2045
Sandai D, Tabana YM, Ouweini AE et al (2016) Resistance of Candida albicans biofilms to drugs and the host immune system. Jundishapur J Microbiol. https://doi.org/10.5812/jjm.37385
Author information
Authors and Affiliations
Contributions
Conception and design of the research: JX and XS; acquisition of data: JX and XS; analysis and interpretation of data: JX and XS; statistical analysis: JX and XS; obtaining funding: JX; drafting the manuscript: XS; revision of manuscript for important intellectual content: JX. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Ethical Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
284_2021_2654_MOESM1_ESM.tif
Supplementary file1 (TIF 17301 KB) Size of the empty vector and complementary strain by PCR. M, DL2000 marker; 1, PCR product of empty vector (size 207 bp); 2, 3, 4, 5, and 6, PCR product of complementary strain (size: 924 bp)
Rights and permissions
About this article
Cite this article
Sun, X., Xiang, J. Mechanism Underlying the Role of LuxR Family Transcriptional Regulator abaR in Biofilm Formation by Acinetobacter baumannii. Curr Microbiol 78, 3936–3944 (2021). https://doi.org/10.1007/s00284-021-02654-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-021-02654-y