Abstract
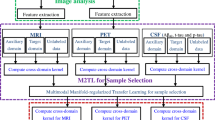
In the early stage of Alzheimer’s disease (AD), mild cognitive impairment (MCI) has a higher risk of progression to AD, so the prediction of whether an MCI subject will progress to AD (known as progressive MCI, PMCI) or not (known as stable MCI, SMCI) within a certain period is particularly important in practice. It is known that such a task could benefit from jointly learning-related auxiliary tasks such as differentiating AD from PMCI or PMCI from normal control (NC) in order to take full advantage of their shared commonality. However, few existing methods along this line fully consider the correlations between the target and auxiliary tasks according to the clinical practice of AD pathology for diagnosis. To deal with this problem, in this paper, treating each task domain as a different one, we borrow the idea from transfer learning and propose a novel multi-auxiliary domain transfer learning (MaDTL) method, which explicitly utilizes the correlations between the target domain (task) and multi-auxiliary domains (tasks) according to the clinical practice. Specifically, the proposed MaDTL method incorporates two key modules. The first one is a multi-auxiliary domain transfer-based feature selection (MaDTFS) model, which can select a discriminative feature subset shared by the target domain and the multi-auxiliary domains. In the MaDTFS model, to combine more training data from multi-auxiliary domains and simultaneously suppress the negative effects resulting from the irrelevant parts of multi-auxiliary domains, we proposed a sparse group correlation Lasso that includes a proposed group correlation Lasso penalty (i.e., \({\Vert \mathbf{W}\mathbf{H}\Vert }_{\mathrm{2,1}}\)) and a proposed correlation Lasso penalty (i.e., \({\Vert \mathbf{W}\mathbf{H}\Vert }_{\mathrm{1,1}}\)). The second module in MaDTL is a multi-auxiliary domain transfer-based classification (MaDTC) model that improves the voting with linear weighting-based ensemble learning. This model extends the constraints of the linear weighting method so that it can simultaneously combine training data from multi-auxiliary domains and achieve a robust classifier by minimizing negative effects from the irrelevant part of multi-auxiliary domains. Experimental results on 409 subjects from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) database with the baseline magnetic resonance imaging (MRI) and cerebrospinal fluid (CSF) data validate the effectiveness of the proposed method by significantly improving the classification accuracy to 80.37% for the identification of MCI-to-AD conversion, outperforming the state-of-the-art methods.









Similar content being viewed by others
Data availability
The datasets used in this paper are from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) which are available at http://adni.loni.usc.edu/. Source code and binary programs developed in this paper are available via email, cb729@nuaa.edu.cn.
Notes
\(\Omega =\{0.0001, 0.0005, 0.0009, 0.001:0.001:0.009, 0.01:0.01:0.09, 0.1:0.1:2\}\)
References
Association A s, (2019). 2019 Alzheimer's disease facts and figures. Alzheimer's & Dement 15, 321–387.
Cheng B, Liu M, Shen D, Zhang D (2019) Robust multi-label transfer feature learning for early diagnosis of Alzheimer’s disease. Brain Imaging Behav 13:138–153
Wee CY, Liu C, Lee A, Joann SP, Ji H, Qiu A (2019) Cortical graph neural network for AD and MCI diagnosis and transfer learning across populations. NeuroImage Clin 23:101929
Choia H, Jinb KH (2018) Predicting cognitive decline with deep learning of brain metabolism and amyloid imaging. Behav Brain Res 344:103–109
Liu X, Goncalves AR, Cao P, Zhao D, Banerjee A (2018) Modeling Alzheimer’s disease cognitive scores using multi-task sparse group lasso. Comput Med Imaging Graph 66:100–114
Zhou K, He W, Xu Y, Xiong G, Cai J (2018) Feature selection and transfer learning for Alzheimer’s disease clinical diagnosis. Appl Sci 8:1372
Hojjati SH, Ebrahimzadeh A, Khazaee A, Feremi AB (2017) Predicting conversion from MCI to AD using resting-state fMRI, graph theoretical approach and SVM. J Neurosci Methods 282:69–80
Kooi T, Litjens G, van Ginneken B, Gubern-Merida A, Sanchez CI, Mann R, den Heeten A, Karssemeijer N (2017) Large scale deep learning for computer aided detection of mammographic lesions. Med Image Anal 35:303–312
Li Q, Wu X, Xu L, Chen K, Yao L, Li R (2017) Multi-modal discriminative dictionary learning for Alzheimer’s disease and mild cognitive impairment. Comput Methods Programs Biomed 150:1–8
Suk HI, Lee SW, Shen D (2017) Deep ensemble learning of sparse regression models for brain disease diagnosis. Med Image Anal 37:101–113
Zhu X, Suk H, Wang L, Lee SW, Shen D (2017) A novel relational regularization feature selection method for joint regression and classification in AD diagnosis. Med Image Anal 38:205–214
Shi B, Chen Y, Zhang P, Smith CD, Liu J (2017) Nonlinear feature transformation and deep fusion for Alzheimer’s disease staging analysis. Pattern Recogn 63:487–498
Cheng B, Liu M, Shen D, Li Z, Zhang D, Alzheimer’s Disease Neuroimaging I (2017) Multi-domain transfer learning for early diagnosis of Alzheimer’s disease. Neuroinformatics 15:115–132
Zhang D, Wang Y, Zhou L, Yuan H, Shen D, ADNI, (2011) Multimodal classification of Alzheimer’s disease and mild cognitive impairment. Neuroimage 55:856–867
Chetelat G, Landeau B, Eustache F, Mezenge F, Viader F, de la Sayette V, Desgranges B, Baron JC (2005) Using voxel-based morphometry to map the structural changes associated with rapid conversion in MCI: a longitudinal MRI study. Neuroimage 27:934–946
Chao LL, Buckley ST, Kornak J, Schuff N, Madison C, Yaffe K, Miller BL, Kramer JH, Weiner MW (2010) ASL perfusion MRI predicts cognitive decline and conversion from MCI to dementia. Alzheimer Dis Assoc Disord 24:19–27
deToledo-Morrell L, Stoub TR, Bulgakova M, Wilson RS, Bennett DA, Leurgans S, Wuu J, Turner DA (2004) MRI-derived entorhinal volume is a good predictor of conversion from MCI to AD. Neurobiol Aging 25:1197–1203
Risacher SL, Saykin AJ, West JD, Shen L, Firpi HA, McDonald BC (2009) Baseline MRI predictors of conversion from MCI to probable AD in the ADNI cohort. Curr Alzheimer Res 6:347–361
Misra C, Fan Y, Davatzikos C (2009) Baseline and longitudinal patterns of brain atrophy in MCI patients, and their use in prediction of short-term conversion to AD: results from ADNI. Neuroimage 44:1415–1422
Liu M, Zhang D, Shen D (2016) Relationship induced multi-template learning for diagnosis of Alzheimer’s disease and mild cognitive impairment. IEEE Trans Med Imaging 35:1463–1474
Bouwman FH, Schoonenboom SNM, van der Flier WM, van Elk EJ, Kok A, Barkhof F, Blankenstein MA, Scheltens P (2007) CSF biomarkers and medial temporal lobe atrophy predict dementia in mild cognitive impairment. Neurobiol Aging 28:1070–1074
Vemuri P, Wiste HJ, Weigand SD, Shaw LM, Trojanowski JQ, Weiner MW, Knopman DS, Petersen RC, Jack CR, ADNI, (2009) MRI and CSF biomarkers in normal, MCI, and AD subjects predicting future clinical change. Neurology 73:294–301
Vemuri P, Wiste HJ, Weigand SD, Shaw LM, Trojanowski JQ, Weiner MW, Knopman DS, Petersen RC, Jack CR, ADNI, (2009) MRI and CSF biomarkers in normal, MCI, and AD subjects diagnostic discrimination and cognitive correlations. Neurology 73:287–293
Lehmann M, Koedam E L, Barnes J, Bartlett J W, Barkhof F, Wattjes M P, Schott J M, Scheltens P, Fox N C, (2012). Visual ratings of atrophy in MCI: prediction of conversion and relationship with CSF biomarkers. Neurobiology of Aging.
Davatzikos C, Bhatt P, Shaw LM, Batmanghelich KN, Trojanowski JQ (2011) Prediction of MCI to AD conversion, via MRI, CSF biomarkers, and pattern classification. Neurobiol Aging 32:2322.e19-2322.e27
Hinrichs C, Singh V, Xu GF, Johnson SC, Neuroimaging AD (2011) Predictive markers for AD in a multi-modality framework: an analysis of MCI progression in the ADNI population. Neuroimage 55:574–589
Liu F, Wee CY, Chen HF, Shen DG, ADNI, (2014) Inter-modality relationship constrained multi-modality multi-task feature selection for Alzheimer’s disease and mild cognitive impairment identification. Neuroimage 84:466–475
Liu M, Zhang J, Yap PT, Shen D (2017) View-aligned hypergraph learning for Alzheimer’s disease diagnosis with incomplete multi-modality data. Med Image Anal 36:123–134
Jie B, Zhang D, Cheng B, Shen D (2015) Manifold regularized multitask feature learning for multimodality disease classification. Hum Brain Mapp 36:489–507
Cheng B, Liu M, Zhang D, Munsell BC, Shen D (2015) Domain transfer learning for MCI conversion prediction. IEEE Trans Biomed Eng 62:1805–1817
Shi J, Zheng X, Li Y, Zhang Q, Ying S (2018) Multimodal neuroimaging feature learning with multimodal stacked deep polynomial networks for diagnosis of Alzheimer’s disease. IEEE J Biomed Health Inform 22:173–183
Liu M, Zhang J, Adeli E, Shen D (2017) Deep multi-task multi-channel learning for joint classification and regression of brain status. MICCAI 2017(3):3–11
Suk H, Lee SW, D. S, ADNI, (2014) Hierarchical feature representation and multimodal fusion with deep learning for AD/MCI diagnosis. Neuroimage 101:569–582
van der Burgh HK, Schmidt R, Westeneng HJ, de Reus MA, van den Berg LH, van den Heuvel MP (2017) Deep learning predictions of survival based on MRI in amyotrophic lateral sclerosis. Neuroimage Clin 13:361–369
Xu J, Luo X, Wang G, Gilmore H, Madabhushi A (2016) A deep convolutional neural network for segmenting and classifying epithelial and stromal regions in histopathological images. Neurocomputing 191:214–223
Zhou J, Liu J, Narayan VA, Ye J, ADNI, (2013) Modeling disease progression via multi-task learning. Neuroimage 78:233–248
Zhang D, Shen D, ADNI, (2012). Multi-modal multi-task learning for joint prediction of multiple regression and classification variables in Alzheimer's disease. NeuroImage 59:895-907
Ye J, Farnum M, Yang E, Verbeeck R, Lobanov V, Raghavan N, Novak G, DiBernardo A, Narayan V A, ADNI, (2012). Sparse learning and stability selection for predicting MCI to AD conversion using baseline ADNI data. Bmc Neurology 12, 1471–2377–12–46.
Zhu X, Suk H, Shen D (2014) A novel matrix-similarity based loss function for joint regression and classification in AD diagnosis. Neuroimage 100:91–105
Wachinger C, Reuter M, ADNI, (2016) Domain adaptation for Alzheimer’s disease diagnostics. NeuroImage 139:470–479
Simon N, Friedman J, Hastie T, Tibshirani R (2013) A sparse-group lasso. J Comput Graph Stat 22:231–245
Chen X, Pan W, Kwok J T, Carbonell J G, (2009). Accelerated gradient method for multi-task sparse learning problem. Proceeding of Ninth IEEE International Conference on Data Mining and Knowledge Discovery, 746–751.
Nemirovski A, (2005). Efficient methods in convex programming.
Argyriou A, Evgeniou T, Pontil M (2008) Convex multi-task feature learning. Mach Learn 73:243–272
Pan SJ, Yang Q (2010) A survey on transfer learning. IEEE Trans Knowl Data Eng 22:1345–1359
Tan B, Song Y, Zhong E, Yang Q, 2015. Transitive transfer learning. the 21th ACM SIGKDD International Conference. ACM.
Tibshirani RJ (1996) Regression shrinkage and selection via the LASSO. J Roy Stat Soc B 58:267–288
DeLong ER, DeLong DM, Clarke-Pearson DL (1988) Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 44:837–845
Eskildsen SF, Coupé P, García-Lorenzo D, Fonov V, Pruessner JC, Collins DL (2013) Prediction of Alzheimer’s disease in subjects with mild cognitive impairment from the ADNI cohort using patterns of cortical thinning. Neuroimage 65:511–521
Cuingnet R, Gerardin E, Tessieras J, Auzias G, Lehericy S, Habert MO, Chupin M, Benali H, Colliot O (2011) Automatic classification of patients with Alzheimer’s disease from structural MRI: a comparison of ten methods using the ADNI database. Neuroimage 56:766–781
Cho Y, Seong JK, Jeong Y, Shin SY, ADNI, (2012) Individual subject classification for Alzheimer’s disease based on incremental learning using a spatial frequency representation of cortical thickness data. Neuroimage 59:2217–2230
Wolz R, Julkunen V, Koikkalainen J, Niskanen E, Zhang DP, Rueckert D, Soininen H, Lotjonen J (2011) Multi-method analysis of MRI images in early diagnostics of Alzheimer’s disease. Plos One 6:e25446
Querbes O, Aubry F, Pariente J, Lotterie J-A, Demonet J-F, Duret V, Puel M, Berry I, Fort J-C, Celsis P, ADNI, (2009) Early diagnosis of Alzheimer’s disease using cortical thickness: impact of cognitive reserve. Brain : a journal of neurology 132:2036–2047
Wee CY, Yap PT, Shen DG, ADNI, (2013) Prediction of Alzheimer’s disease and mild cognitive impairment using cortical morphological patterns. Hum Brain Mapp 34:3411–3425
National Institutes of Health Center for Information Technology (CIT) (2020) Medical image processing, analysis and visualization (MIPAV Version 10.0.0). https://mipav.cit.nih.gov/clickwrap.php
Kabani N, MacDonald D, Holmes CJ, Evans A (1998) A 3D atlas of the human brain. Neuroimage 7:S717
Wang Y, Nie J, Yap P T, Shi F, Guo L, Shen D, (2011). Deformable surface based skull-stripping for large-scale studies. in Medical Image Computing and Computer-Assisted Intervention 3, 635–642.
Zhang YY, Brady M, Smith S (2001) Segmentation of brain MR images through a hidden Markov random field model and the expectation-maximization algorithm. IEEE Trans Med Imaging 20:45–57
Sled JG, Zijdenbos AP, Evans AC (1998) A nonparametric method for automatic correction of intensity nonuniformity in MRI data. IEEE Trans Med Imaging 17:87–97
Shen DG, Davatzikos C (2002) HAMMER: hierarchical attribute matching mechanism for elastic registration. IEEE Trans Med Imaging 21:1421–1439
Acknowledgements
ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: Abbott, AstraZeneca AB, Bayer Schering Pharma AG, Bristol-Myers Squibb, Eisai Global Clinical Development, Elan Corporation, Genentech, GE Healthcare, GlaxoSmithKline, Innogenetics, Johnson and Johnson, Eli Lilly and Co., Medpace, Inc., Merck and Co., Inc., Novartis AG, Pfizer Inc., F. Hoffmann-La Roche, Schering-Plough, Synarc, Inc., as well as nonprofit partners the Alzheimer’s Association and Alzheimer’s Drug Discovery Foundation, with participation from the US Food and Drug Administration. Private sector contributions to ADNI are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory of Neuroimaging at the University of California, Los Angeles.
Funding
Data collection and sharing for this project were funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904). This work was supported by the National Natural Science Foundation of China (No. 61602072), the Chongqing Cutting-edge and Applied Foundation Research Program (No. cstc2018jcyjAX0502), the Scientific and Technological Research Program of Chongqing Municipal Education Commission (No. KJQN202001222), and the Chongqing Three Gorges University Research Program (No.19QN08).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics approval
None.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Cheng, B., Zhu, B. & Pu, S. Multi-auxiliary domain transfer learning for diagnosis of MCI conversion. Neurol Sci 43, 1721–1739 (2022). https://doi.org/10.1007/s10072-021-05568-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10072-021-05568-6