Abstract
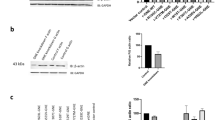
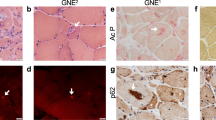
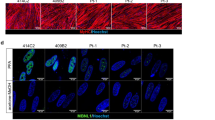
UDP-N-Acetyl glucosamine-2 epimerase/N-acetyl mannosamine kinase (GNE) catalyzes key enzymatic reactions in the biosynthesis of sialic acid. Mutation in GNE gene causes GNE myopathy (GNEM) characterized by adult-onset muscle weakness and degeneration. However, recent studies propose alternate roles of GNE in other cellular processes beside sialic acid biosynthesis, particularly interaction of GNE with α-actinin 1 and 2. Lack of appropriate model system limits drug and treatment options for GNEM as GNE knockout was found to be embryonically lethal. In the present study, we have generated L6 rat skeletal muscle myoblast cell-based model system carrying one single Gne allele where GNE gene is knocked out at exon-3 using AAV mediated SEPT homology recombination (SKM-GNEHz). The cell line was heterozygous for GNE gene with one wild type and one truncated allele as confirmed by sequencing. The phenotype showed reduced GNE epimerase activity with little reduction in sialic acid content. In addition, the heterozygous GNE knockout cells revealed altered cytoskeletal organization with disrupted actin filament. Further, we observed increased levels of RhoA leading to reduced cofilin activity and causing reduced F-actin polymerization. The disturbed signaling cascade resulted in reduced migration of SKM-GNEHz cells. Our study indicates possible role of GNE in regulating actin dynamics and cell migration of skeletal muscle cell. The skeletal muscle cell-based system offers great potential in understanding pathomechanism and target identification for GNEM.








Similar content being viewed by others
Data Availability
All the data are enclosed in the manuscript and supplementary materials. Anything else required shall be provided.
Code Availability
NA
Abbreviations
- GNE:
-
UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase
- ER:
-
Endoplasmic reticulum
- AAV:
-
Adeno-associated viruses
- SEPT:
-
Synthetic exon promoter trap
- ERK:
-
Extracellular signal-regulated kinase
- FAK:
-
Focal adhesion kinase
- Src:
-
Proto-oncogene tyrosine-protein kinase (sarcoma)
- ManNac:
-
N-acetylmannosamine
- NCAM:
-
Neural cell adhesion molecule
- UPR:
-
Unfolded protein response
- CRMP1:
-
Collapsin response mediator protein 1
- ROCK:
-
Rho-associated protein kinase
- LIMK:
-
LIM (Lin-11 Isl-1 Mec-3) kinase
- Aβ 1–42:
-
Amyloid-β 1–42
- loxP:
-
Locus of X-over P1
References
Nestler-parr S, Korchagina D, Toumi M et al (2018) Challenges in research and health technology assessment of rare disease technologies : report of the ISPOR Rare Disease Special Interest Group. Value Heal 21:493–500. https://doi.org/10.1016/j.jval.2018.03.004
Awasthi K, Arya R, Bhattacharya A, Bhattacharya S (2019) The inherited neuromuscular disorder GNE myopathy: research to patient care. Neurol India 67:1213–1219. https://doi.org/10.4103/0028-3886.271259
Devi S, Yadav R, Chanana P, Arya R (2018) Fighting the cause of Alzheimer’s and GNE myopathy. Front Neurosci 12:1–14. https://doi.org/10.3389/fnins.2018.00669
Nonaka I, Sunohara N, Ishiura S, Satoyoshi E (1981) Familial distal myopathy with rimmed vacuole and lamellar (myeloid) body formation. J Neurol Sci 51:141–155. https://doi.org/10.1016/0022-510X(81)90067-8
Argov Z, Yarom R (1984) “Rimmed vacuole myopathy” sparing the quadriceps. A unique disorder in Iranian Jews. J Neurol Sci 64:33–43. https://doi.org/10.1016/0022-510X(84)90053-4
Nishino I, Noguchi S, Murayama K et al (2002) Distal myopathy with rimmed vacuoles is allelic to hereditary inclusion body myopathy. Neurology. https://doi.org/10.1212/01.WNL.0000041631.28557.C6
Huizing M, Carrillo-Carrasco N, Malicdan MCV et al (2014) GNE myopathy: new name and new mutation nomenclature. Neuromuscul Disord. https://doi.org/10.1016/j.nmd.2014.03.004
Huizing M, Krasnewich DM (2009) Hereditary inclusion body myopathy: a decade of progress. Biochim Biophys Acta - Mol Basis Dis 1792:881–887
Eisenberg I, Avidan N, Potikha T et al (2001) The UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase gene is mutated in recessive hereditary inclusion body myopathy. Nat Genet 29:83–87. https://doi.org/10.1038/ng718
Carrillo N, Malicdan MC, Huizing M (2018) GNE Myopathy: etiology, diagnosis, and therapeutic challenges. Neurotherapeutics 15:900–914. https://doi.org/10.1007/s13311-018-0671-y
Benyamini H, Kling Y, Yakovlev L et al (2020) Upregulation of hallmark muscle genes protects GneM743T/M743T mutated knock-in mice from kidney and muscle phenotype. J Neuromuscul Dis. https://doi.org/10.3233/JND-190461
Pogoryelova O, Wilson IJ, Mansbach H et al (2019) GNE genotype explains 20% of phenotypic variability in GNE myopathy. Neurol Genet. https://doi.org/10.1212/NXG.0000000000000308
Nalini A, Gayathri N, Dawn R (2010) Distal myopathy with rimmed vacuoles: report on clinical characteristics in 23 cases. Neurol India 58:235–241. https://doi.org/10.4103/0028-3886.63804
Nishino I, Carrillo-Carrasco N, Argov Z (2015) GNE myopathy: current update and future therapy. J Neurol Neurosurg Psychiatry 86:385–392
Pogoryelova O, González Coraspe JA, Nikolenko N et al (2018) GNE myopathy: From clinics and genetics to pathology and research strategies. Orphanet J Rare Dis 13:1–15. https://doi.org/10.1186/s13023-018-0802-x
Celeste FV, Vilboux T, Ciccone C et al (2014) Mutation update for GNE gene variants associated with GNE myopathy. Hum Mutat 35:915–926. https://doi.org/10.1002/humu.22583
Nalini A, Nishino I, Gayathri N, Hayashi Y (2013) GNE myopathy in India. Neurol India. https://doi.org/10.4103/0028-3886.117609
Chamova T, Guergueltcheva V, Gospodinova M, et al (2015) GNE myopathy in Roma patients homozygous for the p.I618T founder mutation. Neuromuscul Disord. https://doi.org/10.1016/j.nmd.2015.07.004
Haghighi A, Nafissi S, Qurashi A et al (2016) Genetics of GNE myopathy in the non-Jewish Persian population. Eur J Hum Genet. https://doi.org/10.1038/ejhg.2015.78
Chakravorty S, Berger K, Arafat D et al (2019) Clinical utility of RNA sequencing to resolve unusual GNE myopathy with a novel promoter deletion. Muscle Nerve. https://doi.org/10.1002/mus.26486
Hinderlich S, Weidemann W, Yardeni T et al (2015) UDP-GlcNAc 2-epimerase/ManNAc kinase (GNE): a master regulator of sialic acid synthesis. Top Curr Chem. https://doi.org/10.1007/128_2013_464
Yonekawa T, Malicdan MCV, Cho A et al (2014) Sialyllactose ameliorates myopathic phenotypes in symptomatic GNE myopathy model mice. Brain 137:2670–2679. https://doi.org/10.1093/brain/awu210
Argov Z, Caraco Y, Lau H et al (2016) Aceneuramic acid extended release administration maintains upper limb muscle strength in a 48-week study of subjects with GNE myopathy: results from a phase 2, randomized, controlled study. J Neuromuscul Dis. https://doi.org/10.3233/JND-159900
Lochmüller H, Behin A, Caraco Y et al (2019) A phase 3 randomized study evaluating sialic acid extended-release for GNE myopathy. Neurology. https://doi.org/10.1212/WNL.0000000000006932
Valles-Ayoub Y, Esfandiarifard S, Sinai P et al (2012) Serum neural cell adhesion molecule is hyposialylated in hereditary inclusion body myopathy. Genet Test Mol Biomarkers 16:313–317. https://doi.org/10.1089/gtmb.2011.0146
Grover S, Arya R (2014) Role of UDP-N-Acetylglucosamine2-epimerase/N-acetylmannosamine kinase (GNE) in β1-integrin-mediated cell adhesion. Mol Neurobiol. https://doi.org/10.1007/s12035-013-8604-6
Chanana P, Padhy G, Bhargava K, Arya R (2017) Mutation in GNE downregulates peroxiredoxin IV altering ER redox homeostasis. NeuroMolecular Med. https://doi.org/10.1007/s12017-017-8467-5
Sela I, Yakovlev L, Becker Cohen M et al (2013) Variable phenotypes of knockin mice carrying the M712T Gne mutation. NeuroMolecular Med. https://doi.org/10.1007/s12017-012-8209-7
Li H, Chen Q, Liu F et al (2013) Unfolded protein response and activated degradative pathways regulation in GNE myopathy. PLoS One 8.https://doi.org/10.1371/journal.pone.0058116
Sela I, Krentsis IM, Shlomai Z et al (2011) The proteomic profile of hereditary inclusion body myopathy. PLoS ONE. https://doi.org/10.1371/journal.pone.0016334
Weidemann W, Stelzl U, Lisewski U et al (2006) The collapsin response mediator protein 1 (CRMP-1) and the promyelocytic leukemia zinc finger protein (PLZF) bind to UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase (GNE), the key enzyme of sialic acid biosynthesis. FEBS Lett 580:6649–6654. https://doi.org/10.1016/j.febslet.2006.11.015
Amsili S, Zer H, Hinderlich S et al (2008) UDP-N-acetylglucosamine-2-epimerase/N-acetylmannosamine kinase (GNE) binds to alpha-actinin 1: novel pathways in skeletal muscle? PLoS One 3.https://doi.org/10.1371/journal.pone.0002477
Harazi A, Becker-Cohen M, Zer H et al (2017) The Interaction of UDP-N-Acetylglucosamine 2-Epimerase/N-Acetylmannosamine Kinase (GNE) and Alpha-Actinin 2 Is Altered in GNE Myopathy M743T Mutant. Mol Neurobiol 54:2928–2938. https://doi.org/10.1007/s12035-016-9862-x
Henderson CA, Gomez CG, Novak SM et al (2017) Overview of the muscle cytoskeleton. Compr Physiol. https://doi.org/10.1002/cphy.c160033
Kowalski K, Kołodziejczyk A, Sikorska M et al (2017) Stem cells migration during skeletal muscle regeneration - the role of Sdf-1/Cxcr4 and Sdf-1/Cxcr7 axis. Cell Adhes Migr. https://doi.org/10.1080/19336918.2016.1227911
Kim JH, Jin P, Duan R, Chen EH (2015) Mechanisms of myoblast fusion during muscle development. Curr Opin Genet Dev 32:162–170. https://doi.org/10.1016/j.gde.2015.03.006
Fan J, Chan C, McNamara EL et al (2018) Molecular Consequences of the Myopathy-Related D286G Mutation on Actin Function. Front Physiol. https://doi.org/10.3389/fphys.2018.01756
Horak I, Rohde E, Wiechens N et al (2002) Sialylation is essential for early development in mice. Proc Natl Acad Sci. https://doi.org/10.1073/pnas.072066199
Schwarzkopf M, Knobeloch K-P, Rohde E et al (2002) Sialylation is essential for early development in mice. Proc Natl Acad Sci. https://doi.org/10.1073/pnas.072066199
Galeano B, Klootwijk R, Manoli I et al (2007) Mutation in the key enzyme of sialic acid biosynthesis causes severe glomerular proteinuria and is rescued by N-acetylmannosamine. J Clin Invest 117:1585–1594. https://doi.org/10.1172/JCI30954
Malicdan MCV, Noguchi S, Nishino I (2007) Autophagy in a mouse model of distal myopathy with rimmed vacuoles or hereditary inclusion body myopathy. Autophagy 3:396–398. https://doi.org/10.1177/1756285609359546
Daya A, Vatine GD, Becker-Cohen M et al (2014) Gne depletion during zebrafish development impairs skeletal muscle structure and function. Hum Mol Genet. https://doi.org/10.1093/hmg/ddu045
Bosch-Morató M, Iriondo C, Guivernau B, et al (2016) Increased amyloid β-peptide uptake in skeletal muscle is induced by hyposialylation and may account for apoptosis in GNE myopathy. Oncotarget 7:13354–71. https://doi.org/10.18632/oncotarget.7997
Tanner ME (2005) The enzymes of sialic acid biosynthesis. Bioorg Chem 33:216–228. https://doi.org/10.1016/j.bioorg.2005.01.005
Rago C, Vogelstein B, Bunz F (2007) Genetic knockouts and knockins in human somatic cells. Nat Protoc. https://doi.org/10.1038/nprot.2007.408
Van Helvert S, Storm C, Friedl P (2017) Mechanoreciprocity in cell migration. Nat Cell Biol 20:8–20. https://doi.org/10.1038/s41556-017-0012-0
DesMarais V, Ghosh M, Eddy R, Condeelis J (2004) Cofilin takes the lead. J Cell Sci. https://doi.org/10.1242/jcs.01631
Suetsugu S, Takenawa T (2003) Regulation of cortical actin networks in cell migration. Int Rev Cytol. https://doi.org/10.1016/S0074-7696(03)29006-9
Ono S (2010) Dynamic regulation of sarcomeric actin filaments in striated muscle. Cytoskeleton 67:677–692. https://doi.org/10.1002/cm.20476
Sampath, S. C., Sampath, S. C., & Millay, D. P. (2018). Myoblast fusion confusion: the resolution begins. Skelet.Muscle. 8(1). https://doi.org/10.1186/s13395-017-0149-3
Webster MT, Fan CM (2013) c-MET regulates myoblast motility and myocyte fusion during adult skeletal muscle regeneration. PLoS ONE. https://doi.org/10.1371/journal.pone.0081757
Nobes CD, Hall A (1999) Rho GTPases control polarity, protrusion, and adhesion during cell movement. J Cell Biol. https://doi.org/10.1083/jcb.144.6.1235
Mu X, Usas A, Tang Y et al (2013) RhoA mediates defective stem cell function and heterotopic ossification in dystrophic muscle of mice. FASEB J. https://doi.org/10.1096/fj.13-233460
Costa P, Scales TME, Ivaska J, Parsons M (2013) Integrin-Specific Control of Focal Adhesion Kinase and RhoA Regulates Membrane Protrusion and Invasion. PLoS One 8.https://doi.org/10.1371/journal.pone.0074659
Katoh K, Kano Y, Noda Y (2010) Rho-associated kinase-dependent contraction of stress fibres and the organization of focal adhesions. J R Soc Interface 8:305–311. https://doi.org/10.1098/rsif.2010.0419
Destaing O, Sanjay A, Itzstein C et al (2008) The tyrosine kinase activity of c-Src regulates actin dynamics and organization of podosomes in osteoclasts. Mol Biol Cell. https://doi.org/10.1091/mbc.E07-03-0227
Graham ZA, Gallagher PM, Cardozo CP (2015) Focal adhesion kinase and its role in Graham, Z. A., Gallagher, P. M., & Cardozo, C. P. (2015). Focal adhesion kinase and its role in skeletal muscle. J Muscle Res Cell Motil 36:305–315. https://doi.org/10.1007/s10974-015-9415-3
Li SY, Mruk DD, Cheng CY (2013) Focal adhesion kinase is a regulator of F-actin dynamics. Spermatogenesis. https://doi.org/10.4161/spmg.25385
Tsai WC, Yu TY, Lin LP et al (2017) Platelet rich plasma promotes skeletal muscle cell migration in association with up-regulation of FAK, paxillin, and F-Actin formation. J Orthop Res. https://doi.org/10.1002/jor.23547
Pollard TD, Cooper JA (2009) Actin, a Central Player in Cell Shape and Movement. Science 326:1208–1212. https://doi.org/10.1126/science.1175862
Chen SC, Huang CH, Lai SJ et al (2016) Mechanism and inhibition of human UDP-GlcNAc 2-epimerase, the key enzyme in sialic acid biosynthesis. Sci Rep. https://doi.org/10.1038/srep23274
Noguchi S, Keira Y, Murayama K et al (2004) Reduction of UDP-N-acetylglucosamine 2-epimerase/N-acetylmannosamine kinase activity and sialylation in distal myopathy with rimmed vacuoles. J Biol Chem 279:11402–11407. https://doi.org/10.1074/jbc.M313171200
Devi SS, Yadav R, Arya R (2021) Altered actin dynamics in cell migration of GNE mutant cells 9:1–18. https://doi.org/10.3389/fcell.2021.603742
Singh R, Arya R (2016) GNE myopathy and cell apoptosis: a comparative mutation analysis. Mol Neurobiol 53:3088–3101. https://doi.org/10.1007/s12035-015-9191-5
Singh R, Chaudhary P, Arya R (2018) Role of IGF-1R in ameliorating apoptosis of GNE deficient cells. Sci Rep. https://doi.org/10.1038/s41598-018-25510-9
Amsili S, Zer H, Hinderlich S et al (2008) UDP-N-acetylglucosamine-2-epimerase/N-acetylmannosamine kinase (GNE) binds to alpha-actinin 1: novel pathways in skeletal muscle? PLoS ONE. https://doi.org/10.1371/journal.pone.0002477
Harazi A, Becker-Cohen M, Zer H et al (2017) The Interaction of UDP-N-acetylglucosamine 2-epimerase/N-Acetylmannosamine kinase (GNE) and alpha-actinin 2 is altered in GNE myopathy M743T mutant. Mol Neurobiol. https://doi.org/10.1007/s12035-016-9862-x
Castellani L, Salvati E, Alemà S, Falcone G (2006) Fine regulation of RhoA and Rock is required for skeletal muscle differentiation. J Biol Chem. https://doi.org/10.1074/jbc.M601390200
Huveneers S, Danen EHJ (2009) Adhesion signaling - crosstalk between integrins, Src and Rho. J Cell Sci. https://doi.org/10.1242/jcs.039446
Urciuolo A, Urbani L, Perin S et al (2018) Decellularised skeletal muscles allow functional muscle regeneration by promoting host cell migration. Sci Rep. https://doi.org/10.1038/s41598-018-26371-y
Wang W, Chen M, Gao Y et al (2018) P2Y6 regulates cytoskeleton reorganization and cell migration of C2C12 myoblasts via ROCK pathway. J Cell Biochem. https://doi.org/10.1002/jcb.26350
Ridley AJ (2011) Life at the Leading Edge. Cell 145:1012–1022. https://doi.org/10.1016/j.cell.2011.06.010
Ambriz X, de Lanerolle P, Ambrosio JR (2018) The mechanobiology of the actin cytoskeleton in stem cells during differentiation and interaction with biomaterials. Stem Cells Int 1–11. https://doi.org/10.1155/2018/2891957
Babij P, Booth FW (1988) α-Actin and cytochrome c mRNAs in atrophied adult rat skeletal muscle. Am J Physiol - Cell Physiol 254.https://doi.org/10.1152/ajpcell.1988.254.5.c651
Holinstat M, Knezevic N, Broman M et al (2006) Suppression of RhoA activity by focal adhesion kinase-induced activation of p190RhoGAP: role in regulation of endothelial permeability. J Biol Chem. https://doi.org/10.1074/jbc.M511248200
Pirone DM, Liu WF, Ruiz SA et al (2006) An inhibitory role for FAK in regulating proliferation: a link between limited adhesion and RhoA-ROCK signaling. J Cell Biol. https://doi.org/10.1083/jcb.200510062
Yang W, Hu P (2018) Skeletal muscle regeneration is modulated by inflammation. J Orthop Transl 13:25–32. https://doi.org/10.1016/j.jot.2018.01.002
Lehka L, Rędowicz MJ (2020) Mechanisms regulating myoblast fusion: A multilevel interplay. Semin Cell Dev Biol 104:81–92. https://doi.org/10.1016/j.semcdb.2020.02.004
Lepper C, Conway SJ, Fan C (2010) Distinct genetic requirements 460:627–631. https://doi.org/10.1038/nature08209.Adult
Blanchoin L, Boujemaa-Paterski R, Sykes C, Plastino J (2014) Actin dynamics, architecture, and mechanics in cell motility. Physiol Rev. https://doi.org/10.1152/physrev.00018.2013
Karolczak J, Pavlyk I, Majewski Ł et al (2015) Involvement of unconventional myosin VI in myoblast function and myotube formation. Histochem Cell Biol. https://doi.org/10.1007/s00418-015-1322-6
Yuan Z, Qiao C, Hu P et al (2011) A versatile adeno-associated virus vector producer cell line method for scalable vector production of different serotypes. Hum Gene Ther. https://doi.org/10.1089/hum.2010.241
Bork K, Reutter W, Weidemann W, Horstkorte R (2007) Enhanced sialylation of EPO by overexpression of UDP-GlcNAc 2-epimerase/ManAc kinase containing a sialuria mutation in CHO cells. FEBS Lett. https://doi.org/10.1016/j.febslet.2007.07.060
Blume A, Ghaderi D, Liebich V et al (2004) UDP-N-acetylglucosamine2-epimerase/N-acetylmannosamine kinase, functionally expressed in and purified from Escherichia coli, yeast, and insect cells. Protein Expr Purif 35:387–396. https://doi.org/10.1016/j.pep.2004.02.013
Reissig JL, Strominger JL, Leloir LF (1955) A modified colorimetric method for the estimation of N-acetyl amino sugars. J Biol Chem 217:959–966. https://doi.org/10.1016/s0021-9258(18)65959-9
Acknowledgements
We acknowledge Prof. Sudha Bhattacharya, School of Environmental Sciences, Jawaharlal Nehru University, India, and Prof. Alok Bhattacharya, Ashoka University, for their support and help during the project. We also acknowledge Dr. Sivaprakash Ramalingam, IGIB, for valuable suggestions. We acknowledge Advanced Instrumentation Research Facility, JNU, and its staff members (Ashok Sahu and Prabhat Yadav) for helping in acquiring confocal images.
Funding
This work was supported by:
1. Science and Engineering Research Board, Department of Science and Technology, Ministry of Science and Technology, Government of India, New Delhi; Grant Number-EMR/2015/001798.
2. Department of Atomic Energy, Government of India; Grant Number-37(1)/14/31/2018-BRNS/37253
Author information
Authors and Affiliations
Contributions
RA conceptualized, designed, brought financial support through grants, and analyzed the data. RA compiled and wrote the manuscript. SSD performed the experiments, generated the cell line and characterized the cell line, and helped in manuscript draft writing. RY validated the data by performing experiments, blind study and editing of the manuscript. FM confirmed the cell lines through sequencing and performed sialic acid content study. PC characterized the cell line by performing epimerase assay and helped in literature search. SS helped in editing of the manuscript. All the authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics Approval
The study was performed in the laboratory cell lines and does not involve human or animal subjects.
Consent to Participate
NA.
Consent for Publication
All the authors have given consent for publication.
Conflict of Interest
The authors declare no conflict of interest.
Competing Interest Statement
The current work is related to a patent application number 202011019014 dated on 4 May 2020 in the Indian Patent Office, titled 'A skeletal Muscle cell-based model for GNE Myopathy'. This association does not alter the author adherence to all Molecular Neurobiology policies on sharing data as detailed online.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Devi, S.S., Yadav, R., Mashangva, F. et al. Generation and Characterization of a Skeletal Muscle Cell-Based Model Carrying One Single Gne Allele: Implications in Actin Dynamics. Mol Neurobiol 58, 6316–6334 (2021). https://doi.org/10.1007/s12035-021-02549-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-021-02549-w