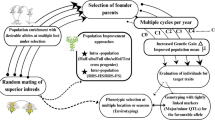
Abstract
Phenotypic selection, for accumulating the desirable characters to develop the crops tailored as per our need, has been the basis of the plant breeding since the time immemorial. But with the several issues associated with this approach viz., huge time, resource and space intensive approach, researchers are on continuous journey to explore more efficient approach with high throughput that could substantially deduct the time and resource requirement with high fidelity. With the advent of several molecular markers, newer avenue of the crop improvement has been explored and marker-assisted recurrent selection stand out to be an intensive approach, which has provided the breeder an efficient tool to use his skills more rigorously. Shortening the time while enhancing the selection efficiency and, thus improving the overall genetic gain has made this approach the need of the hour and choice of the breeders. This review takes you to the journey of its advent and its potential role in population improvement in both self- and cross-pollinated crops.




Similar content being viewed by others
References
Abasht B, Sandford E, Arango J, Settar P, Fulton JE, O’Sullivan NP, Hassen A, Habier D, Fernando RL, Dekkers JCM, Lamont SL (2009) Extent and consistency of linkage disequilibrium and identification of DNA markers for production and egg quality traits in commercial layer chicken populations. BMC Genom 10:S2
Abdallah AA, Al AM, Geiger HH, Parzies HK (2009) Marker-assisted recurrent selection for increased out crossing in Caudatum–race Sorghum. In Proceedings of the international conference on applied biotechnology, pp 28–30
Abdulmalik RO, Menkir A, Meseka SK, Unachukwu N, Ado SG, Olarewaju JD, Gedil M (2017) Genetic gains in grain yield of a maize population improved through marker assisted recurrent selection under stress and non-stress conditions in West Africa. Front Plant Sci 8:841
Albrecht T, Auinger HJ, Wimmer V, Ogutu JO, Knaak C, Ouzunova M, Schön CC (2014) Genome-based prediction of maize hybrid performance across genetic groups, testers, locations, and years. Theor Appl Genet 127:1375–1386
Alexandrov N, Tai S, Wang W, Mansueto L, Palis K, Fuentes RR, Ulat VJ, Chebotarov D, Zhang G, Li Z, Mauleon R, Hamilton RS, McNally KL (2015) SNP-seek database of SNPs derived from 3000 rice genomes. Nucleic Acids Res 43:D1023–D1027
Araus JL, Kefauver SC, Zaman-Allah M, Olsen MS, Cairns JE (2018) Translating high throughput phenotyping into genetic gain. Trends Plant Sci 23:451–466
Arora L, Narula A (2017) Gene editing and crop improvement using CRISPR-Cas9 system. Front Plant Sci 8:1932
Bankole F, Menkir A, Olaoye G, Crossa J, Hearne S, Unachukwu N, Gedil M (2017) Genetic gains in yield and yield related traits under drought stress and favorable environments in a maize population improved using marker assisted recurrent selection. Front Plant Sci 8:808
Beckmann JS, Soller M (1986) Restriction fragment length polymorphisms and genetic improvement of agricultural species. Euphytica 35:111–124
Berilli APCG, Pereira MG, Gonçalves LSA, da Cunha KS, Ramos HCC, do Souza FilhoAmaral Júnior GA (2011) Use of molecular markers in reciprocal recurrent selection of maize increases heterosis effects. Genet Mol Res 10:2589–2596
Beyene Y, Semagn K, Crossa J, Mugo S, Atlin GN, Tarekegne A, Bänziger M (2015) Improving maize grain yield under drought stress and non-stress environments in sub-Saharan Africa using marker-assisted recurrent selection. Crop Sci 56:344–353
Beyene Y, Semagn K, Mugo S, Prasanna BM, Tarekegne A, Gakunga J, Crossa J (2016) Performance and grain yield stability of maize populations developed using marker-assisted recurrent selection and pedigree selection procedures. Euphytica 208:285–297
Bhattramakki D, Rafalski A (2001) Discovery and application of single nucleotide polymorphism markers in plants. Plant genotyping: The DNA fingerprinting of plants, pp 179–192
Bimpong IK, Manneh B, El-Namkay R et al (2014) Mapping QTLs related to salt tolerance in rice at the young seedling stage using 384-plex single nucleotide polymorphism SNP, marker sets. Mol Plant Breed 5:47–63
Boyle EA, Li YI, Pritchard J (2017) A expanded view of complex traits: from polygenic to omnigenic. Cell 169:1177–1186
Burow MD, Blake TK (1998) Molecular tools for the study of complex traits. In Molecular dissection of complex traits, pp 13–29
Cerrudo D, Cao S, Yuan Y, Martinez C, Suarez EA, Babu R, Trachsel S (2018) Genomic selection outperforms marker assisted selection for grain yield and physiological traits in a maize doubled haploid population across water treatments. Front Plant Sci 9:366
Charmet G, Robert N, Perretant MR, Gay G, Sourdille P, Groos C, Bernard M (1999) Marker-assisted recurrent selection for cumulating additive and interactive QTLs in recombinant inbred lines. Theor Appl Genet 99:1143–1148
Charmet G, Robert N, Perretant MR, Gay G, Sourdille P, Groos C, Bernard M (2001) Marker assisted recurrent selection for cumulating QTLs for bread-making related traits. Euphytica 119:89–93
Choudhary A, Kumar A, Kaur N (2020a) ROS and oxidative burst: roots in plant development. Plant Divers 42(1):33–43
Choudhary A, Kumar A, Kaur N, Paul A (2020b) Plasmodesmata the nano bridges in plant cell: are the answer for all the developmental processes? Russian J Plant Physiol 67:785–796
Choudhary A, Kumar A, Kaur H, Kaur N (2021) MiRNA: the taskmaster of plant world. Biologia 76:1551–1567
Collard BCY, Mackill DJ (2008) Marker-assisted selection: an approach for precision plant breeding in the 21st century. Philos Trans R Soc B 363:557–572
Collard BCY, Beredo JC, Lenaerts B, Mendoza R, Santelices R, Lopena V, Verdeprado H, Raghavan C, Gregorio GB, Vial L, Demont M, Biswas PS, Iftekharuddaula KM, Rahman MA, Cobb JN, Islam MR (2017) Revisiting rice breeding methods: evaluating the use of rapid generation advance (RGA) for routine rice breeding. Plant Prod Sci 20:337–352
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Zhang F, Zhang F (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339:819–823
Crosbie TM, Eathington SR, Johnson Sr GR, Edwards M, Reiter R, Stark S, Lamkey KR (2006) Plant breeding: past, present, and future. In Plant breeding: the Arnel R. Hallauer international symposium. Ames, Iowa, USA: Blackwell Publishing, pp 3–50
Doi K, Iwata N, Yoshimura A (1997) The construction of chromosome substitution lines of African rice (Oryza glaberrima Steud.) in the background of Japonica rice (O sativa L.). Rice Genet News 14:39–41
Dudley JW, Moll RH (1969) Interpretation and use of estimates of heritability and genetic variances in plant breeding. Crop Sci 9:257–262
Eathington SR, Crosbie TM, Edwards MD, Reiter RS, Bull JK (2007) Molecular markers in a commercial breeding program. Crop Sci 47:154
Edwards M, Johnson L (1994) RFLPs for rapid recurrent selection. Analysis of molecular marker data. Joint Plant Breeding Symposia Series American Society of Agronomy, Madison, WI, pp 33–40
Falconer D, Mackay TTFC (1996) Quantitative genetics
Falconer DS, Mackay TFC, Frankham R (1996) Introduction to quantitative genetics, 4th edn London. UK: Benjamin Cummings
Garrido-Cardenas JA, Mesa-Valle C, Manzano-Agugliaro F (2018) Trends in plant research using molecular markers. Planta 247:543–557
Gilliham M, Able JA, Roy SJ (2017) Translating knowledge about abiotic stress tolerance to breeding programmes. Plant J 90:898–917
Habash DZ, Kehel Z, Nachit M (2009) Genomic approaches for designing durum wheat ready for climate change with a focus on drought. J Exp Bot 60:2805–2815
Habier D, Fernando RL, Dekkers JCM (2009) Genomic selection using low-density marker panels. Genetics 182:343–353
Han B, Wang C, Tang Z, Ren Y, Li Y, Zhang D, Dong Y, Zhao X (2015) Genome-wide analysis of microsatellite markers based on sequenced database in Chinese spring wheat (Triticum aestivum L.). PLoS One 10:e0141540
Heffner EL, Jannink J, Sorrells ME (2011) Genomic selection accuracy using multifamily prediction models in a wheat breeding program. Plant Genome 4:65–75
Jiang GL (2013) Molecular markers and marker-assisted breeding in plants. Plant breeding from laboratories to fields, pp 45–83
Jinek M, East A, Cheng A, Lin S, Ma E, Doudna J (2013) RNA-programmed genome editing in human cells. eLife 2:10
Johnson R (2004) Marker assisted selection. In: Jannick J (ed) Plant Breed Rev 24:293–309
Kim SM, Suh JP, Lee CK, Le JH, Kim YG, Jena KK (2014) QTL mapping and development of candidate gene-derived DNA markers associated with seedling cold tolerance in rice (Oryza sativa L.). Mol Genet Genom 289:333–343
Kubo T, Aida Y, Nakamura K, Tsunematsu H, Doi K, Yoshimura A (2002) Reciprocal chromosome segment substitution series derived from japonica and indica cross of rice (Oryza sativa L.). Breed Sci 52:319–325
Kumar A, Sharma A, Sharma R, Srivastva P, Choudhary A (2021a) Exploration of wheat wild relative diversity from Lahaul valley: a cold arid desert of Indian Himalayas. Cereal Res Commun. https://doi.org/10.1007/s42976-021-00166-w
Kumar A, Sharma A, Sharma R, Choudhary A, Srivastava P, Kaur H, Padhy AK (2021b) Morpho-physiological evaluation of Elymus semicostatus (Nees ex Steud.) Melderis as potential donor for drought tolerance in Wheat (Triticum aestivum L.). Genen Resour Crop Evol. https://doi.org/10.1007/s10722-021-01241-1
Kumar A, Choudhary A, Kaur N, Kaur H (2021c) Wake Up: it’s Time to Bloom. Russian J Plant Physiol 68:579–595
Lande R, Thompson R (1990) Efficiency of marker-assisted selection in the improvement of quantitative traits. Genetics 124:743–756
Lee M (1995) DNA markers and plant breeding programs. Adv Agron 55:265–344
Lee SY, Ahn JH, Cha YS, Yun DW, Lee MC, Ko JC, Lee KS, Eun MY (2007) Mapping QTLs related to salinity tolerance of rice at the young seedling stage. Plant Breed 126:43–46
Li C (2020) Breeding crops by design for future agriculture. J Zhejiang Univ Sci B 21:423
Li S, Zhao B, Yuan D, Duan M, Qian Q, Tang L et al (2013) Rice zinc finger protein dst enhances grain production through controlling gn1a/osckx2 expression. Proc Natl Acad Sci USA 110:3167–3172
Lu K, Wei L, Li X, Wang Y, Wu J et al (2019) Whole-genome resequencing reveals Brassica napus origin and genetic loci involved in its improvement. Nat Commun 10:1154
Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE et al (2013) RNA-guided human genome engineering via Cas9. Science 339(6121):823–826
Marziliano N, Notarangelo MF, Cereda M, Caporale V, Coppini L, Demola MA, Guidorossi A, Crocamo A, Pigazzani F, Boffetti F, del Giudice F, Orsini F, Pirola D, Cocci A, Manzalini C, Casu G, Bianchessi M, Ardissino D, Merlini PA (2015) Rapid and portable, lab-on-chip, point-of-care genotyping for evaluating clopidogrel metabolism. Clin Chim Acta 451:240–246
Massman JM, Jung HJG, Bernardo R (2013) Genomewide selection versus marker assisted recurrent selection to improve grain yield and Stover quality traits for cellulosic ethanol in maize. Crop Sci 53:58–66
Mayor PJ, Bernardo R (2009) Genomewide selection and marker assisted recurrent selection in doubled haploid versus F2 populations. Crop Sci 49:1719–1725
Michel S, Ametz C, Gungor H, Akgöl B, Epure D, Grausgruber H, Buerstmayr H (2017) Genomic assisted selection for enhancing line breeding: merging genomic and phenotypic selection in winter wheat breeding programs with preliminary yield trials. Theor Appl Genet 130:363–376
Muthamilarasan M, Prasad M (2015) Advances in Setaria genomics for genetic improvement of cereals and bioenergy grasses. Theor Appl Genet 128:1–14
Pandey G, Misra G, Kumari K, Gupta S, Parida SK, Chattopadhyay D, Prasad M (2013) Genome-wide development and use of microsatellite markers for large-scale genotyping applications in foxtail millet [Setaria italica (L.)]. DNA Res 20:197–207
Paulo M-J, Boer M, Huang X, Koornneef M, van Eeuwijk F (2008) A mixed model QTL analysis for a complex cross population consisting of a half diallel of two-way hybrids in Arabidopsis thaliana: analysis of simulated data. Euphytica 161:107–114
Peleman JD, Van der Voort JR (2003) Breeding by design. Trends Plant Sci 8:330–334
Pingali PL (2012) Green revolution: impacts, limits, and the path ahead. Proc Natl Acad Sci USA 109:12302–12308
Prasanna BM (2016) Developing and deploying abiotic stress-tolerant maize varieties in the tropics: challenges and opportunities. Molecular Breeding for Sustainable Crop Improvement. Springer, Cham, pp 61–77
Rahman MA, Bimpong IK, Bizimana JB, Pascual ED, Arceta M, Swamy BPM et al (2017) Mapping QTLs using a novel source of salinity tolerance from Hasawi and their interaction with environments in rice. Rice 10:47
Rahman M, Davies P, Bansal U, Pasam R, Hayden M, Trethowan R (2020) Marker-assisted recurrent selection improves the crown rot resistance of bread wheat. Mol 40:1–14
Ribaut JM, De Vicente MC, Delannay X (2010) Molecular breeding in developing countries: challenges and perspectives. Curr Opin Plant Biol 13:213–218
RibautJ M, Edmeades G, Perotti E, Hoisington D (2000) QTL analyses, MAS results, and perspectives for drought-tolerance improvement in tropical maize. Prod Water-Limited Environ. 131
Schwantes IA, do Amaral AT, de Almeida Filho JE, Vivas M, Cabral PDS, Guimarães AG, Ferreira FRA (2018) First Report of Recurrent Genomic Selection with Real Data in Popcorn and Genetic Gain Increases. BioRxiv, 466003
Singh BD (2012) Plant breeding, principles and methods, 9th edn. Kalyani Publishers, New Delhi
Singh M, Choudhary A, Kumar A, Thapa S (2021) Genetically modified food: their problems and promise. Biotic Res Today 3(2):111–113
Sorrells ME (2015) Genomic selection in plants: empirical results and implications for wheat and barley breeding programs. In: Ogihara Y et al (eds) Advances in wheat genetics: from genome to field.
Stam P (1994) Marker assisted breeding. JW van Ooijen, J. Jansen (eds.), In Biometrics in plant breeding: applications of molecular markers, . Proc. 9th meeting Eucarpia Section Biometrics in Plant Breeding. CPRO-DLO, Wageningen, pp 32–44
Sun Z, Yin X, Ding J, Yu D, Hu M, Sun X et al (2017) QTL analysis and dissection of panicle components in rice using advanced backcross populations derived from Oryza sativa cultivars HR1128 and Nipponbare. PLoS ONE 12:e0175692
Suryendra PJ, Revathi P, Singh AK, Viraktamath BC (2020) Marker assisted recurrent selection for genetic male sterile population improvement in rice. Electron J Plant Breed 11:149–155
Sweeney MT, Thomson MJ, Pfeil BE, McCouch S (2006) Caught red-handed: Rc encodes a basic helix-loop-helix protein conditioning red pericarp in rice. Plant Cell 18:283–294
Thapa S, Baral R, Thapa S (2019) Status, Challenges and Solutions of Oil-Seed Production in India. Res Rev J Agri Allied Sci 8(1):27–34
Thomson MJ, Tai TH, McClung AM, Lai X-H, Hing ME, Lobos KB, Xu Y, Martinez CP, McCouch SR (2003) Mapping quantitative trait loci for yield, yield components and morphological traits in an advanced backcross population between Oryza rufipogon and the Oryza sativa cultivar Jefferson. Theor Appl Genet 107:479–493
Tsaih SW, Lu L, Airey DC, Williams RW, Churchill GA (2005) Quantitative trait mapping in a diallel cross of recombinant inbred lines. Mamm Genome 16:344–355
Wang J, Eagles HA, Trethowan R, Van Ginkel M (2005) Using computer simulation of the selection process and known gene information to assist in parental selection in wheat quality breeding. Aust J Agric Res 56:465–473
Wang S, Wong D, Forrest K, Allen A et al (2014a) Characterization of polyploid wheat genomic diversity using a high-density 90,000 single nucleotide polymorphism array. Plant Biotechnol J 12:787–796
Wang X, Jiang GL, Green M, Scott RA, Hyten DL, Cregan PB (2014b) Quantitative trait locus analysis of unsaturated fatty acids in a recombinant inbred population of soybean. Mol Genet Genom 33:281–296
Wang X, Jiang GL, Green M, Scott RA, Song Q, Hyten DL, Cregan PB (2014c) Identification and validation of quantitative trait loci for seed yield, oil and protein contents in two recombinant inbred line populations of soybean. Mol Genet Genom 289:935–949
Woolliams JA, Berg P, Dagnachew BS, Meuwissen THE (2015) Genetic contributions and their optimization. J Anim Breed Genet 132:89–99
Xu J, Yuan Y, Xu Y, Zhang G, Guo X, Wu F, Wang Q, Rong T, Pan G, Cao M, Tang Q, Gao S, Liu Y, Wang J, Lan H, Lu Y (2014) Identification of candidate genes for drought tolerance by wholegenome resequencing in maize. BMC Plant Biol 14:83
Yi CX, Guo WZ, Zhu XF, Min LF, Zhang TZ (2004) Pyramiding breeding by marker assisted recurrent selection in upland cotton II: selection effects on resistance to Helicoverpa armigera. Sci Sin 37:801–807
Zhou G, Zhang Q, Tan C, Zhang XQ, Li C (2015) Development of genome-wide InDel markers and their integration with SSR, DArT and SNP markers in single barley map. BMC Genom 16:804
Zhou Y, Dong G, Tao Y, Chen C, Yang B, Wu Y et al (2016) Mapping quantitative trait loci associated with toot traits using sequencingbased genotyping chromosome segment substitution lines derived from 9311 and Nipponbare in Rice (Oryza sativa L.). PLoS ONE 11(3):e0151796
Zhu Z, Tan L, Fu Y, Liu F, Cai H, Xie D, Wu F, Wu J, Matsumoto T, Sun C (2013) Genetic control of inflorescence architecture during rice domestication. Nat Commun 4:2200
Author information
Authors and Affiliations
Contributions
All the authors have equal contribution in writing a manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by A. Goyal.
Rights and permissions
About this article
Cite this article
Singh, M., Nara, U., Kumar, A. et al. Enhancing genetic gains through marker-assisted recurrent selection: from phenotyping to genotyping. CEREAL RESEARCH COMMUNICATIONS 50, 523–538 (2022). https://doi.org/10.1007/s42976-021-00207-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42976-021-00207-4