Advances in the Development of Microbial Double-Stranded RNA Production Systems for Application of RNA Interference in Agricultural Pest Control
- 1State Key Laboratory of Wheat and Maize Crop Science/College of Plant Protection, Henan Agricultural University, Zhengzhou, China
- 2Key Laboratory of Insect Developmental and Evolutionary Biology, CAS Center for Excellence in Molecular Plant Sciences, Shanghai Institute of Plant Physiology and Ecology, Innovation Academy for Seed Design, Chinese Academy of Sciences, Shanghai, China
RNA interference (RNAi) is a valuable and revolutionary technology that has been widely applied in medicine and agriculture. The application of RNAi in various industries requires large amounts of low-cost double-stranded RNA (dsRNA). Chemical synthesis can only produce short dsRNAs; long dsRNAs need to be synthesized biologically. Several microbial chassis cells, such as Escherichia coli, Saccharomyces cerevisiae, and Bacillus species, have been used for dsRNA synthesis. However, the titer, rate of production, and yield of dsRNA obtained by these microorganism-based strategies is still low. In this review, we summarize advances in microbial dsRNA production, and analyze the merits and faults of different microbial dsRNA production systems. This review provides a guide for dsRNA production system selection. Future development of efficient microbial dsRNA production systems is also discussed.
Introduction
The large-scale use of chemical pesticides creates tremendous ecological pressure on soil, water, air, and the human living environment. After the long-term use of chemical pesticides, resistance, resurgence, and residue (3R) problems have become increasingly prominent (Tudi et al., 2021). The emergence of RNA interference (RNAi) technology has brought new hope of solving these problems. In this technology, double-stranded RNA (dsRNA) enters a host and triggers the RNAi effect—the expression of the complementary target gene is silenced, which affects the growth and development of the target organism, thus achieving pest control (Fire et al., 1998; Fletcher et al., 2020; Zhu and Palli, 2020). RNAi pesticides are considered novel, and ecofriendly, because RNAi technology uses precise targeting and the pesticide agent could be easily degradable.
However, several problems need to be solved before this technology can be widely applied, such as efficient, high-throughput target gene acquisition, dsRNA delivery strategies in different organisms (insects, plants, fungi, bacteria, and viruses), the stability of the dsRNA in field application, and construction of multi-species integrated control strategies in complex ecological environments (Zhang et al., 2013). Besides, large-scale, low-cost synthesis of dsRNA is crucial for applying RNAi technology in agriculture (Silver et al., 2021). Chemical synthesis of RNA is suitable for the synthesis of short RNAs, such as small interfering RNAs (siRNAs), because the synthesis error rate increases and the yield decreases when the length of the target RNA product increases (Mu et al., 2018). In vitro synthesis strategies relying on T7/SP6 RNA polymerase and in vivo synthesis by engineered bacteria are often used for dsRNA synthesis. The in vitro synthesis strategy can produce high-purity dsRNA, but the cost is relatively high. Moreover, this method requires auxiliary materials, such as DNA templates, enzymes, and nucleotides (Mu et al., 2018). The in vivo synthesis strategy produces low-cost dsRNA in high yields, but this strategy requires later purification of the product and inactivation of the engineered microbial strain (Mendiola et al., 2020). Nevertheless, the in vivo synthesis strategy is more likely to reduce dsRNA production costs and increase yields in the future (Cooper et al., 2021). In this review, applications of microbe-mediated dsRNA expression systems are summarized, and the selection of efficient microbial dsRNA production systems is discussed.
Escherichia coli dsRNA Expression Systems
E. coli is a commonly used bacterium for dsRNA expression because of its clear genetic background and convenient genetic manipulation. E. coli strain HT115 (DE3), which is RNase III deficient, and L4440 vector with a pair of oppositely oriented T7 promoters (one on each side of the multiple cloning site) are widely used as an expression strain and vector for dsRNA production (Timmons et al., 2001; Voloudakis et al., 2015). After introduction of the L4440 vector ligated with the target fragment into strain HT115 (DE3), large amounts of T7 RNA polymerase can be synthesized on induction by isopropyl β-D-1-thiogalactopyranoside (IPTG); the T7 RNA polymerase binds to the T7 promoter in L4440, which mediates the transcription of downstream DNA sequences into RNA. As a result, two complementary RNAs are synthesized, which in turn form the target dsRNA (Voloudakis et al., 2015).
The production of dsRNA using engineered bacterial expression was first attempted by Timmons and Fire (1998), and the corresponding RNAi phenotype was verified after feeding to the nematode Caenorhabditis elegans, showing that dsRNA expressed by bacteria can induce RNAi effects (Timmons et al., 2001). Using this dsRNA generation strategy, RNAi effects were induced in a variety of insects, such as Spodoptera exigua (Tian et al., 2009), Bactrocera dorsalis (Li et al., 2011), Chilo infuscatellus (Zhang et al., 2012), Spodoptera exigua (Vatanparast and Kim, 2017), Plagiodera versicolora (Zhang et al., 2019), Tuta absoluta (Bento et al., 2020), Harmonia axyridis (Ma et al., 2020), Spodoptera littoralis (Caccia et al., 2020), Agrilus planipennis (Leelesh and Rieske, 2020). In addition, expressed virus dsRNA can protect a plant or animal against viral infection. For example, E. coli strain HT115 (DE3) was used to express dsRNA of the Chinese Sacbrood Virus (CSBV) VP1, which was fed to Chinese honeybees (Apis cerana) and effectively prevented the virus infecting the bees (Zhang et al., 2016). Treating Nicotiana benthamiana with dsRNAs of fragments of two major plant viruses, Pepper Mild Mottle Virus (PMMoV) and Plum Pox Virus (PPV), effectively reduced the infection of N. benthamiana by these two viruses (Tenllado et al., 2003). All these results show that engineered E. coli can synthesize dsRNAs, and the dsRNA produced can induce RNAi effects in the corresponding target organisms.
The yield of dsRNA synthesized in E. coli has been improved over time. An average of 4 μg of dsRNA was obtained per ml of E. coli culture in 2003 (Tenllado et al., 2003), and 45 μg hairpin dsRNA per ml of bacteria (optical density at 600 nm = 1) in 2013 (Posiri et al., 2013). The increase of dsRNA yield is due to the fermentation methods and operation parameters used (Thammasorn et al., 2015; Papic et al., 2018). dsRNA production using batch fermentation and fed-batch fermentation was compared in a 10 L fermenter, and the dsRNA titer in fed-batch fermentation (95.0 ± 21.5 μg/ml) was nearly 30-fold that found in batch fermentation (3.4 ± 0.5 μg/ml) (Thammasorn et al., 2015). The nutrition can also affect the final dsRNA yield, and the production of dsRNA using Terrific broth (TB) (6.2 ± 0.2 μg/ml) was higher than that using Luria-Bertani (LB) broth (2.6 ± 0.8 μg/ml). After further optimization, the yield was close to 0.06 g/g, the maximum production rate reached 11.1 mg L−1 h−1 by batch fermentation and 15.2 mg L−1 h−1 by fed-batch fermentation (Papic et al., 2018). Therefore, the dsRNA yield is related to bacterial growth, and fed-batch fermentation resulted in a higher dsRNA yield by sustainably supplying nutrition.
Modification of the expression vector and host strain can further improve the efficiency of dsRNA synthesis. dsRNA production using a new E. coli expression system, pET28-BL21 (DE3) RNase III- was thrice than that of L4440-HT115 (DE3) (Ma et al., 2020).
Moreover, extraction methods are closely linked to the yield of dsRNA. The titer of dsRNA extracted from E. coli by ultrasonic crushing and phenol extraction was 19.5 μg/ml, while sonication and heating before dsRNA extraction increased the titer of dsRNA by 2.5- to 5- fold (Ahn et al., 2019).
Nowadays, large-scale synthesis of dsRNA in E. coli has developed, but further increasing the titer, rate, and yield (TRY) of dsRNA production is essential for future applications.
Saccharomyces cerevisiae dsRNA Expression Systems
The model eukaryotic species Saccharomyces cerevisiae has also been used as a chassis for dsRNA production. S. cerevisiae has a clear genetic background, easy genetic engineering methods, and well-developed fermentation processes (Nandy and Srivastava, 2018). Besides, S. cerevisiae does not contain the core genes Dicer-2 and Argonaute-2 of the RNAi pathway (Drinnenberg et al., 2009), which allows efficient dsRNA synthesis in S. cerevisiae compared with E. coli and other bacterial species (Zhong et al., 2019). Similarly, plant chloroplast does not contain RNAi pathway, and dsRNA can be enriched to 0.4% of total RNA in plant chloroplast; expression of dsRNA in plant chloroplast can be used to protect plants from being fed by insects, which would be more efficient than expressing dsRNA form the plant leaves (Zhang et al., 2015).
Feeding the fruit fly Drosophila suzukii with recombinant yeast expressing insect dsRNA targeting y-Tubulin resulted in a significant reduction in larval survivorship, adult motility, and reproduction (Murphy et al., 2016). Moreover, feeding D. suzukii with genetically modified S. cerevisiae expressing dsRNA (targeting y-tubulin23C) resulted in a significant decrease in the fitness of D. suzukii in the environment (Abrieux and Chiu, 2016).
The expression of dsRNA in S. cerevisiae has also been validated in the mosquito Aedes aegypti. Fez2 and lrc were selected as target genes in A. aegypti, and shRNAs of these genes were expressed in S. cerevisiae. When the genetically-modified S. cerevisiae was heated, dried, and fed to insects, this led to >95%mortality of A. aegypti (Hapairai et al., 2017). The same effects were observed with Aedes albopictus, Anopheles gambiae, and Culex quinquefasciatus (Mysore et al., 2017; Mysore et al., 2019a; Mysore et al., 2019b). In this way, biocontrol strategies for specific mosquito species can be developed, to effectively suppress human diseases transmitted by mosquitoes.
Many insects, livestock, aquaculture species and humans consume yeast. Therefore, developing efficient yeast expression systems might increase the possibility of applying yeast-derived dsRNA commercially (Duman-Scheel, 2019). The dsRNA produced by S. cerevisiae can also serve as a potential oral delivery system for shRNA to mammalian cells (mouse intestinal DCs) and be used in human disease therapeutics (Zhang et al., 2014; Duman-Scheel, 2019). Several companies have developed yeast dsRNA expression systems. In May 2019, Renaissance BioScience filed a patent application for the production and delivery of bioactive dsRNA ingredients using yeasts.
In the future, large-scale production of dsRNA in S. cerevisiae can be enhanced by improving the expression vectors (Crook et al., 2014), the promoters for the dsRNA transcription (Voineagu et al., 2008), the length of the hairpins (Yoshimatsu and Nagawa, 1989), and the sites of integration positions (Kim et al., 2015).
Bacillus dsRNA Expression Systems
Some B. subtilis strains are classified as probiotics for human and animal consumption (Rosales-Mendoza and Angulo, 2015). Therefore, this species has also been selected for dsRNA expression. The dsRNA (daf-2, unc-62) expression vector pBSR was introduced into B. subtilis, and feeding C. elegans this genetically-modified B. subtilis strain induced RNAi effects (Lezzerini et al., 2015). A B. subtilis strain with dsVP28 expression was able to effectively prevent shrimp infection with white spot syndrome virus (WSSV); the survival rate of shrimp treated with the B. subtilis strain was 91.67%, while that in the control group was only 28.57% (Saelim et al., 2020).
Bacillus thuringiensis (Bt) is an effective biopesticide production strain that has been widely used for control of lepidopteran pests. Bt has been used as an expression host for dsRNA production. In the vector pBTdsSBV-VP1, two spore-producing-dependent cyt1Aa promoters in opposite direction were linked to the VP1 gene of Sacbrood virus (SBV), and a Shine-Dalgarno sequence (GAAAGGAGG) was added at specific positions, which increased the stability of the RNA. Transfer of pBTdsSBV-VP1 into Bt strain 4Q7 led to the expression of dsRNA. Feeding the total RNA extracted from this Bt strain to Apis cerana (honeybees) infected with SBV virus significantly reduced the viral infection of the insects (Park et al., 2020).
The Bt-based dsRNA production platform has some advantages compared with other platforms. The cry sporulation-dependent gene promotor was used for dsRNA expression, and the dsRNA could be produced during the sporulation phase of Bt. Moreover, other expression systems (like E. coli, B. subtiis, S. cerevisiae expression systems) require an inducer (IPTG or others) to induce dsRNA expression, but no inducer is needed for expression in Bt. Finally, Bt cells can undergo enzyme-associated autolysis after sporulation, thus cell lysis is not required for dsRNA extraction (Park et al., 2020).
With the increase of insect resistance to Bt, the use of Bt as a platform for dsRNA expression would help with pest control via a Bt + RNAi strategy (Caccia et al., 2020; Kang et al., 2021). Therefore, the Bt dsRNA expression system could be a useful dsRNA production platform for the introduction of RNAi in organisms.
Insect-Symbiotic Bacteria dsRNA Expression Systems
There are abundant symbiotic bacteria in the oral tract and gut of insects, and they interact directly with the insects and plants. Some symbiotic bacteria can easily be genetically manipulated, so they might be potentially efficient dsRNA production platforms for insect control. The use of insect-symbiotic bacteria to express dsRNA for insect control is known as symbiont-mediated RNAi (SMR) (Taracena et al., 2015; Whitten et al., 2016; Whitten and Dyson, 2017; Whitten, 2019; Asgari et al., 2020).
Rhodococcus rhodnii (R. rhodnii), a symbiotic bacterium of the triatomine Rhodnius prolixus, was used to express RHBP-specific hairpin RNA; the gene expression products of RHBP can suppress R. prolixus by affecting its adult oviposition (Taracena et al., 2015). Subsequently, two symbiotic bacterial strains, R. rhodnii and BFo2 (a member of the Enterobacteriales), were isolated from the insects R. prolixus and Frankliniella occidentalis (western flower thrips), respectively. The RNase III gene was knocked out and dsRNA expression cassettes was expressed in these two insect symbiotic bacteria; when the engineered bacteria were taken up by insects, the dsRNA functioned in the hosts, inducing RNAi effects (Whitten et al., 2016).
Snodgrassella alvi, a core gut symbiotic bacterium of the honeybee A. mellifera, was modified as a dsRNA-producing host. The dsRNA produced by the engineered S. alvi can suppress gene expression in A. mellifera. Moreover, this dsRNA can suppress genes of parasitic Varroa mites and kill them, which protects the honey-bees from the Varroa mites, the most threatening pest to the world’s beekeeping industry (Leonard et al., 2020). Based on this technology, a new bioproduct, “BioDirect” was registered as dsRNA for the prevention and control of Varroa mites. This is the first dsRNA biopesticide active ingredient submitted to the U.S. Environmental Protection Agency (EPA) for exogenous application in agriculture.
Thus, SMR is not only potential pest control agents, but can also be beneficial for insect protection. SMR depends on both the specificity of RNAi toward the targeted insect gene, and the specificity of the symbiotic bacterium for its host. This dual specificity makes SMR a precision control tool, and this tool is obviously different from chemical insecticides. However, there are issues that need to be addressed before symbiotic bacteria can reliably serve as dsRNA expression hosts. The first is to find suitable symbiotic bacteria that stably colonize the host insects; the bacterial content should also be relatively high in the host insect. The second is that the symbiotic bacteria must be able to express dsRNA efficiently and stably. Thus, the acquisition and modification of symbiotic bacteria and colonization of the engineered symbiotic bacteria in the host need to be addressed before applying SMR for dsRNA production.
Nevertheless, this SMR strategy is specific for pest control without increasing environmental stress, and it might be widely used in the future.
Conclusion and Perspectives
Genetic engineering of microorganisms for large-scale production of dsRNA is feasible. Currently, E. coli, Bacillus, S. cerevisiae and several other symbiotic bacteria have mature expression systems for dsRNA production. As most of these bacteria are probiotics and/or model species, they might be the most suitable microbial hosts for diverse dsRNA production. Corynebacterium glutamicum has also been shown to efficiently synthesize dsRNA longer than 1 kbp in a yield >1 g/L of culture (Hashero et al., 2021). Besides, microalgae can also be engineered as dsRNA expression vectors, and shrimps and crabs can be protected from bacterial or viral infection by feeding on microalgae expressing dsRNA (Saksmerprome et al., 2009; Somchai et al., 2016; Charoonnart et al., 2019). Fungi (Chen et al., 2015) and viruses (Dubreuil et al., 2009; Kumar et al., 2012) have also been engineered to produce dsRNA, and better results have been obtained.
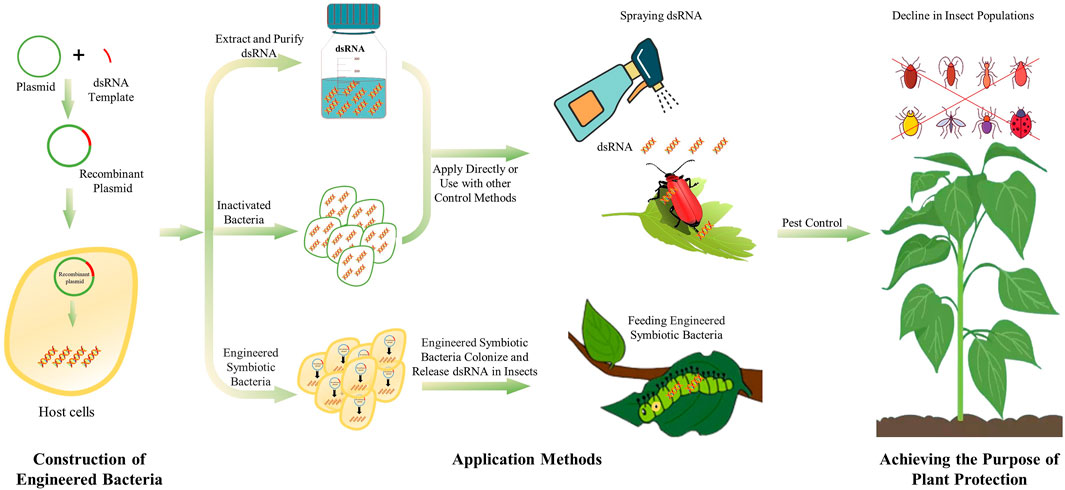
The dsRNA synthesized by microbes can be used directly in live or inactivated microbes. However, engineered microbes entering a host induce immune responses, which might compromise the desired RNAi effects. Moreover, the engineered microbes may spread into the environment, and lead to sustainable expression of dsRNA, which might affect non-target species in the environment. Besides, plasmid-based expression elements may be transferred inter-species, resulting in biological contamination problems (Mendelsohn et al., 2020). dsRNA produced in engineered bacteria cannot be secreted directly outside the cell. Therefore, lysis, extraction and purification are required to obtained dsRNA production. The lysis of cells can be performed by ultrasonication, enzymatic lysis, boiling lysis, while sodium dodecyl sulfate (SDS) can be used to enhance the lysis (Posiri et al., 2013). After the cell wall is broken, the nucleic acid can be released to obtain a crude extract of dsRNA. Then, use appropriate RNA extraction methods, such as TRIzol reagent or other RNA extraction reagents, to obtain pure dsRNA production. Extracting and purifying dsRNA from engineered bacteria will avoid or reduce the problems mentioned above. However, these processes are relatively cumbersome and need to be further optimized. The dsRNA obtained through microbial-production can be directly applied to pest control by spraying, and the nanocarrier-mediated transdermal dsRNA delivery system can facilitate the development of sprayable RNA pesticides (Zheng et al., 2019; Yan et al., 2020). Which method to use also needs to be selected according to different environments (Figure 1).
There are also some further technical issues in this field that need to be solved. For example, substrates for industrial fermentation can be contaminated with various bacteria, and such contaminants can inhibit growth of the desired (dsRNA-expressing) bacteria and reduce the efficiency of the fermentation process, thus significantly reducing productivity (Seo et al., 2020). Antimicrobial decontamination strategies have been developed, but the metabolites produced and antibiotics used to avoid contamination by other microorganisms are released, inevitably putting pressure on the environment and increasing risks to human health (Kraemer et al., 2019). Once these problems are solved and dsRNAs can be produced by large-scale fermentation, they will have broad application prospects and bring huge economic benefits.
dsRNA production methods have been continuously optimized in recent years to adapt to production needs and promote the application of this technology. The cost of dsRNA was approximately US$12,000/g in 2008, dropping to US$60/g in 2018, and in 2020, RNAGri had the ability to produce tons of dsRNA at a cost of US$1/g, Greenlight’s GreenWorX™ system can further reduce the cost of dsRNA synthesis to < US$0.5/g (Cagliari et al., 2019; Suhag et al., 2020; Taning et al., 2020), which will provide material for the economical large-scale application of dsRNA-based pesticides.
The large-scale application of RNAi technology relies on the construction of efficient and appropriate microbial cell factories for dsRNA production. With the development of synthetic biology, global rewiring of the expression systems of model species to increase dsRNA expression levels will be possible. In the future, active engineered microorganisms for dsRNA production and low-cost purified dsRNA will become available leading to greener agriculture without chemical pesticides to protect plants from insects and microbial infections.
Author Contributions
RG: Conceptualization, Writing - Original Draft; DC: Writing - Original Draft; XH: Writing - Original Draft; XM: Writing - Review and Editing; HL: Writing - Review and Editing.
Funding
This work was supported by the Shanghai Agriculture Applied Technology Development Program, China (Grant No. X2019-02-08-00-12-F01125), the National Natural Science Foundation of China (31772520; 31702057), the Educational Commission of Henan Province of China (21A210017), the Young Talents Promotion Project in Henan Province (2020HYTP038), the Science and Technology Planning Project of Henan Province of China (212102110442).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We thank James Allen, DPhil, from Liwen Bianji (Edanz) (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
References
Abrieux, A., and Chiu, J. C. (2016). Oral Delivery of dsRNA by Microbes: Beyond Pest Control. Communicative Integr. Biol. 9, e1236163. doi:10.1080/19420889.2016.1236163
Ahn, S. J., Donahue, K., Koh, Y., Martin, R. R., and Choi, M. Y. (2019). Microbial-Based Double-Stranded RNA Production to Develop Cost-Effective RNA Interference Application for Insect Pest Management. Int. J. Insect Sci. 11, 1179543319840323. doi:10.1177/1179543319840323
Asgari, M., Ilbeigikhamsehnejad, M., Rismani, E., Dinparast Djadid, N., and Raz, A. (2020). Molecular Characterization of RNase III Protein of Asaia Sp. For Developing a Robust RNAi-Based Paratransgensis Tool to Affect the Sexual Life-Cycle of Plasmodium or Anopheles Fitness. Parasites Vectors 13, 42. doi:10.1186/s13071-020-3889-6
Bento, F. M., Marques, R. N., Campana, F. B., Demétrio, C. G., Leandro, R. A., Parra, J. R. P., et al. (2020). Gene Silencing by RNAi via Oral Delivery of dsRNA by Bacteria in the South American Tomato Pinworm, Tuta Absoluta. Pest Manag. Sci. 76, 287–295. doi:10.1002/ps.5513
Caccia, S., Astarita, F., Barra, E., Di Lelio, I., Varricchio, P., and Pennacchio, F. (2020). Enhancement of Bacillus Thuringiensis Toxicity by Feeding Spodoptera Littoralis Larvae with Bacteria Expressing Immune Suppressive dsRNA. J. Pest Sci. 93, 303–314. doi:10.1007/s10340-019-01140-6
Cagliari, D., Dias, N. P., Galdeano, D. M., Dos Santos, E. Á., Smagghe, G., and Zotti, M. J. (2019). Management of Pest Insects and Plant Diseases by Non-Transformative RNAi. Front. Plant Sci. 10, 1319. doi:10.3389/fpls.2019.01319
Charoonnart, P., Worakajit, N., Zedler, J. A. Z., Meetam, M., Robinson, C., and Saksmerprome, V. (2019). Generation of Microalga Chlamydomonas Reinhardtii Expressing Shrimp Antiviral dsRNA without Supplementation of Antibiotics. Sci. Rep. 9, 3164. doi:10.1038/s41598-019-39539-x
Chen, X., Li, L., Hu, Q., Zhang, B., Wu, W., Jin, F., et al. (2015). Expression of dsRNA in Recombinant Isaria Fumosorosea Strain Targets the TLR7 Gene in Bemisia Tabaci. BMC Biotechnol. 15, 64. doi:10.1186/s12896-015-0170-8
Cooper, A. M., Song, H., Yu, Z., Biondi, M., Bai, J., Shi, X., et al. (2021). Comparison of Strategies for Enhancing RNA Interference Efficiency in Ostrinia Nubilalis. Pest Manag. Sci. 77, 635–645. doi:10.1002/ps.6114
Crook, N. C., Schmitz, A. C., and Alper, H. S. (2014). Optimization of a Yeast RNA Interference System for Controlling Gene Expression and Enabling Rapid Metabolic Engineering. ACS Synth. Biol. 3, 307–313. doi:10.1021/sb4001432
Drinnenberg, I. A., Weinberg, D. E., Xie, K. T., Mower, J. P., Wolfe, K. H., Fink, G. R., et al. (2009). RNAi in Budding Yeast. Science 326, 544–550. doi:10.1126/science.1176945
Dubreuil, G., Magliano, M., Dubrana, M. P., Lozano, J., Lecomte, P., Favery, B., et al. (2009). Tobacco Rattle Virus Mediates Gene Silencing in a Plant Parasitic Root-Knot Nematode. J. Exp. Bot. 60, 4041–4050. doi:10.1093/jxb/erp237
Duman-Scheel, M. (2019). Saccharomyces cerevisiae (Baker's Yeast) as an Interfering RNA Expression and Delivery System. Curr. Drug Targets 20, 942–952. doi:10.2174/1389450120666181126123538
Fire, A., Xu, S., Montgomery, M. K., Kostas, S. A., Driver, S. E., and Mello, C. C. (1998). Potent and Specific Genetic Interference by Double-Stranded RNA in Caenorhabditis elegans. Nature 391, 806–811. doi:10.1038/35888
Fletcher, S. J., Reeves, P. T., Hoang, B. T., and Mitter, N. (2020). A Perspective on RNAi-Based Biopesticides. Front. Plant Sci. 11, 51. doi:10.3389/fpls.2020.00051
Hapairai, L. K., Mysore, K., Chen, Y., Harper, E. I., Scheel, M. P., Lesnik, A. M., et al. (2017). Lure-and-Kill Yeast Interfering RNA Larvicides Targeting Neural Genes in the Human Disease Vector Mosquito Aedes aegypti. Sci. Rep. 7, 13223. doi:10.1038/s41598-017-13566-y
Hashiro, S., Chikami, Y., Kawaguchi, H., Krylov, A. A., Niimi, T., and Yasueda, H. (2021). Efficient Production of Long Double-Stranded RNAs Applicable to Agricultural Pest Control by Corynebacterium Glutamicum Equipped with Coliphage T7-Expression System. Appl. Microbiol. Biotechnol. 105, 4987–5000. doi:10.1007/s00253-021-11324-9
Kang, S., Sun, D., Qin, J., Guo, L., Zhu, L., Bai, Y., et al. (2021). Fused: A Promising Molecular Target for an RNAi-Based Strategy to Manage Bt Resistance in Plutella Xylostella (L.). J. Pest Sci. doi:10.1007/s10340-021-01374-3
Kim, H., Yoo, S. J., and Kang, H. A. (2015). Editorial: Yeast Synthetic Biology: New Tools to Unlock Cellular Function. FEMS Yeast Res. 15, 1. doi:10.1093/femsyr/fou003
Kraemer, S. A., Ramachandran, A., and Perron, G. G. (2019). Antibiotic Pollution in the Environment: From Microbial Ecology to Public Policy. Microorganisms 7, 180. doi:10.3390/microorganisms7060180
Kumar, P., Pandit, S. S., and Baldwin, I. T. (2012). Tobacco Rattle Virus Vector: A Rapid and Transient Means of Silencing Manduca Sexta Genes by Plant Mediated RNA Interference. PLoS One 7, e31347. doi:10.1371/journal.pone.0031347
Leelesh, R. S., and Rieske, L. K. (2020). Oral Ingestion of Bacterially Expressed dsRNA Can Silence Genes and Cause Mortality in a Highly Invasive, Tree-Killing Pest, the Emerald Ash Borer. Insects 11, 440. doi:10.3390/insects11070440
Leonard, S. P., Powell, J. E., Perutka, J., Geng, P., Heckmann, L. C., Horak, R. D., et al. (2020). Engineered Symbionts Activate Honey Bee Immunity and Limit Pathogens. Science 367, 573–576. doi:10.1126/science.aax9039
Lezzerini, M., Van De Ven, K., Veerman, M., Brul, S., and Budovskaya, Y. V. (2015). Specific RNA Interference in Caenorhabditis elegans by Ingested dsRNA Expressed in Bacillus Subtilis. PLoS One 10, e0124508. doi:10.1371/journal.pone.0124508
Li, X., Zhang, M., and Zhang, H. (2011). RNA Interference of Four Genes in Adult Bactrocera Dorsalis by Feeding Their dsRNAs. PLoS One 6, e17788. doi:10.1371/journal.pone.0017788
Ma, Z. Z., Zhou, H., Wei, Y. L., Yan, S., and Shen, J. (2020). A Novel Plasmid- Escherichia coli System Produces Large Batch dsRNAs for Insect Gene Silencing. Pest Manag. Sci. 76, 2505–2512. doi:10.1002/ps.5792
Mendelsohn, M. L., Gathmann, A., Kardassi, D., Sachana, M., Hopwood, E. M., Dietz-Pfeilstetter, A., et al. (2020). Summary of Discussions from the 2019 OECD Conference on RNAi Based Pesticides. Front. Plant Sci. 11, 740. doi:10.3389/fpls.2020.00740
Mendiola, S. Y., Civitello, D. J., and Gerardo, N. M. (2020). An Integrative Approach to Symbiont-Mediated Vector Control for Agricultural Pathogens. Curr. Opin. Insect Sci. 39, 57–62. doi:10.1016/j.cois.2020.02.007
Mu, X., Greenwald, E., Ahmad, S., and Hur, S. (2018). An Origin of the Immunogenicity of In Vitro Transcribed RNA. Nucleic Acids Res. 46, 5239–5249. doi:10.1093/nar/gky177
Murphy, K. A., Tabuloc, C. A., Cervantes, K. R., and Chiu, J. C. (2016). Ingestion of Genetically Modified Yeast Symbiont Reduces Fitness of an Insect Pest via RNA Interference. Sci. Rep. 6, 22587. doi:10.1038/srep22587
Mysore, K., Hapairai, L. K., Sun, L., Harper, E. I., Chen, Y., Eggleson, K. K., et al. (2017). Yeast Interfering RNA Larvicides Targeting Neural Genes Induce High Rates of Anopheles Larval Mortality. Malar. J. 16, 461. doi:10.1186/s12936-017-2112-5
Mysore, K., Hapairai, L. K., Wei, N., Realey, J. S., Scheel, N. D., Severson, D. W., et al. (2019a). Preparation and Use of a Yeast shRNA Delivery System for Gene Silencing in Mosquito Larvae. Methods Mol. Biol. 1858, 213–231. doi:10.1007/978-1-4939-8775-7_15
Mysore, K., Li, P., Wang, C.-W., Hapairai, L. K., Scheel, N. D., Realey, J. S., et al. (2019b). Characterization of a Broad-Based Mosquito Yeast Interfering RNA Larvicide with a Conserved Target Site in Mosquito Semaphorin-1a Genes. Parasites Vectors 12, 256. doi:10.1186/s13071-019-3504-x
Nandy, S. K., and Srivastava, R. K. (2018). A Review on Sustainable Yeast Biotechnological Processes and Applications. Microbiol. Res. 207, 83–90. doi:10.1016/j.micres.2017.11.013
Papić, L., Rivas, J., Toledo, S., and Romero, J. (2018). Double-stranded RNA Production and the Kinetics of Recombinant Escherichia coli HT115 in Fed-Batch Culture. Biotechnol. Rep. (Amst) 20, e00292. doi:10.1016/j.btre.2018.e00292
Park, M. G., Kim, W. J., Choi, J. Y., Kim, J. H., Park, D. H., Kim, J. Y., et al. (2020). Development of a Bacillus Thuringiensis Based dsRNA Production Platform to Control Sacbrood Virus in Apis cerana. Pest Manag. Sci. 76, 1699–1704. doi:10.1002/ps.5692
Posiri, P., Ongvarrasopone, C., and Panyim, S. (2013). A Simple One-Step Method for Producing dsRNA from E. coli to Inhibit Shrimp Virus Replication. J. Virol. Methods 188, 64–69. doi:10.1016/j.jviromet.2012.11.033
Rosales-Mendoza, S., and Angulo, C. (2015). Bacillus Subtilis Comes of Age as a Vaccine Production Host and Delivery Vehicle. Expert Rev. Vaccin. 14, 1135–1148. doi:10.1586/14760584.2015.1051469
Saelim, H., Loprasert, S., and Phongdara, A. (2020). Bacillus Subtilis Expressing dsVP28 Improved Shrimp Survival from WSSV challenge. ScienceAsia 46S, 19. doi:10.2306/scienceasia1513-1874.2020.s003
Saksmerprome, V., Charoonnart, P., Gangnonngiw, W., and Withyachumnarnkul, B. (2009). A Novel and Inexpensive Application of RNAi Technology to Protect Shrimp from Viral Disease. J. Virol. Methods 162, 213–217. doi:10.1016/j.jviromet.2009.08.010
Seo, S. O., Park, S. K., Jung, S. C., Ryu, C. M., and Kim, J. S. (2020). Anti-Contamination Strategies for Yeast Fermentations. Microorganisms 8, 274. doi:10.3390/microorganisms8020274
Silver, K., Cooper, A. M., and Zhu, K. Y. (2021). Strategies for Enhancing the Efficiency of RNA Interference in Insects. Pest Manag. Sci. 77, 2645–2658. doi:10.1002/ps.6277
Somchai, P., Jitrakorn, S., Thitamadee, S., Meetam, M., and Saksmerprome, V. (2016). Use of Microalgae Chlamydomonas Reinhardtii for Production of Double-Stranded RNA against Shrimp Virus. Aquacult. Rep. 3, 178–183. doi:10.1016/j.aqrep.2016.03.003
Suhag, A., Yadav, H., Chaudhary, D., Subramanian, S., Jaiwal, R., and Jaiwal, P. K. (2020). Biotechnological Interventions for the Sustainable Management of a Global Pest, Whitefly (Bemisia Tabaci). Insect Sci. doi:10.1111/1744-7917.12853
Taning, C. N., Arpaia, S., Christiaens, O., Dietz‐Pfeilstetter, A., Jones, H., Mezzetti, B., et al. (2020). RNA‐Based Biocontrol Compounds: Current Status and Perspectives to Reach the Market. Pest Manag. Sci. 76, 841–845. doi:10.1002/ps.5686
Taracena, M. L., Oliveira, P. L., Almendares, O., Umaña, C., Lowenberger, C., Dotson, E. M., et al. (2015). Genetically Modifying the Insect Gut Microbiota to Control Chagas Disease Vectors through Systemic RNAi. Plos Negl. Trop. Dis. 9, e0003358. doi:10.1371/journal.pntd.0003358
Tenllado, F., Martínez-García, B., Vargas, M., and Díaz-Ruíz, J. R. (2003). Crude Extracts of Bacterially Expressed dsRNA Can Be Used to Protect Plants against Virus Infections. BMC Biotechnol. 3, 3. doi:10.1186/1472-6750-3-3
Thammasorn, T., Sangsuriya, P., Meemetta, W., Senapin, S., Jitrakorn, S., Rattanarojpong, T., et al. (2015). Large-Scale Production and Antiviral Efficacy of Multi-Target Double-Stranded RNA for the Prevention of white Spot Syndrome Virus (WSSV) in Shrimp. BMC Biotechnol. 15, 110. doi:10.1186/s12896-015-0226-9
Tian, H., Peng, H., Yao, Q., Chen, H., Xie, Q., Tang, B., et al. (2009). Developmental Control of a Lepidopteran Pest Spodoptera Exigua by Ingestion of Bacteria Expressing dsRNA of a Non-Midgut Gene. PLoS One 4, e6225. doi:10.1371/journal.pone.0006225
Timmons, L., Court, D. L., and Fire, A. (2001). Ingestion of Bacterially Expressed dsRNAs Can Produce Specific and Potent Genetic Interference in Caenorhabditis elegans. Gene. 263, 103–112. doi:10.1016/s0378-1119(00)00579-5
Timmons, L., and Fire, A. (1998). Specific Interference by Ingested dsRNA. Nature 395, 854. doi:10.1038/27579
Tudi, M., Daniel Ruan, H., Wang, L., Lyu, J., Sadler, R., Connell, D., et al. (2021). Agriculture Development, Pesticide Application and its Impact on the Environment. Int. J. Environ. Res. Public Health 18, 1112. doi:10.3390/ijerph18031112
Vatanparast, M., and Kim, Y. (2017). Optimization of Recombinant Bacteria Expressing dsRNA to Enhance Insecticidal Activity against a Lepidopteran Insect, Spodoptera Exigua. PLoS One 12, e0183054. doi:10.1371/journal.pone.0183054
Voineagu, I., Narayanan, V., Lobachev, K. S., and Mirkin, S. M. (2008). Replication Stalling at Unstable Inverted Repeats: Interplay between DNA Hairpins and fork Stabilizing Proteins. Proc. Natl. Acad. Sci. 105, 9936–9941. doi:10.1073/pnas.0804510105
Voloudakis, A. E., Holeva, M. C., Sarin, L. P., Bamford, D. H., Vargas, M., Poranen, M. M., et al. (2015). Efficient Double-Stranded RNA Production Methods for Utilization in Plant Virus Control. Methods Mol. Biol. 1236, 255–274. doi:10.1007/978-1-4939-1743-3_19
Whitten, M., and Dyson, P. (2017). Gene Silencing in Non-Model Insects: Overcoming Hurdles Using Symbiotic Bacteria for Trauma-free Sustainable Delivery of RNA Interference: Sustained RNA Interference in Insects Mediated by Symbiotic Bacteria: Applications as a Genetic Tool and as a Biocide. Bioessays 39, 1600247. doi:10.1002/bies.201600247
Whitten, M. M. A., Facey, P. D., Del Sol, R., Fernández-Martínez, L. T., Evans, M. C., Mitchell, J. J., et al. (2016). Symbiont-Mediated RNA Interference in Insects. Proc. R. Soc. B. 283, 20160042. doi:10.1098/rspb.2016.0042
Whitten, M. M. (2019). Novel RNAi Delivery Systems in the Control of Medical and Veterinary Pests. Curr. Opin. Insect Sci. 34, 1–6. doi:10.1016/j.cois.2019.02.001
Yan, S., Qian, J., Cai, C., Ma, Z., Li, J., Yin, M., et al. (2020). Spray Method Application of Transdermal dsRNA Delivery System for Efficient Gene Silencing and Pest Control on Soybean Aphid Aphis Glycines. J. Pest Sci. 93, 449–459. doi:10.1007/s10340-019-01157-x
Yoshimatsu, T., and Nagawa, F. (1989). Control of Gene Expression by Artificial Introns in Saccharomyces cerevisiae. Science 244, 1346–1348. doi:10.1126/science.2544026
Zhang, H., Li, H.-C., and Miao, X.-X. (2013). Feasibility, Limitation and Possible Solutions of RNAi-Based Technology for Insect Pest Control. Insect Sci. 20, 15–30. doi:10.1111/j.1744-7917.2012.01513.x
Zhang, J., Khan, S. A., Hasse, C., Ruf, S., Heckel, D. G., and Bock, R. (2015). Full Crop protection from an Insect Pest by Expression of Long Double-Stranded RNAs in Plastids. Science 347, 991–994. doi:10.1126/science.1261680
Zhang, J., Zhang, Y., and Han, R. (2016). The High-Throughput Production of dsRNA against Sacbrood Virus for Use in the Honey Bee Apis cerana (Hymenoptera: Apidae). Virus Genes 52, 698–705. doi:10.1007/s11262-016-1346-6
Zhang, L., Zhang, T., Wang, L., Shao, S., Chen, Z., and Zhang, Z. (2014). In Vivo targeted Delivery of CD40 shRNA to Mouse Intestinal Dendritic Cells by Oral Administration of Recombinant Sacchromyces Cerevisiae. Gene Ther. 21, 709–714. doi:10.1038/gt.2014.50
Zhang, Y., Xu, L., Li, S., and Zhang, J. (2019). Bacteria-Mediated RNA Interference for Management of Plagiodera Versicolora (Coleoptera: Chrysomelidae). Insects 10, 415. doi:10.3390/insects10120415
Zhang, Y. L., Zhang, S. Z., Kulye, M., Wu, S. R., Yu, N. T., Wang, J. H., et al. (2012). Silencing of Molt-Regulating Transcription Factor Gene, CiHR3, Affects Growth and Development of Sugarcane Stem Borer, Chilo Infuscatellus. J. Insect Sci. 12, 91. doi:10.1673/031.012.9101
Zheng, Y., Hu, Y., Yan, S., Zhou, H., Song, D., Yin, M., et al. (2019). A Polymer/Detergent Formulation Improves dsRNA Penetration through the Body wall and RNAi‐Induced Mortality in the Soybean Aphid Aphis Glycines. Pest Manag. Sci. 75, 1993–1999. doi:10.1002/ps.5313
Zhong, C., Smith, N. A., Zhang, D., Goodfellow, S., Zhang, R., Shan, W., et al. (2019). Full-Length Hairpin RNA Accumulates at High Levels in Yeast but Not in Bacteria and Plants. Genes (Basel) 10, 458. doi:10.3390/genes10060458
Keywords: RNA interference, dsRNA, microbes, synthetic biology, production
Citation: Guan R, Chu D, Han X, Miao X and Li H (2021) Advances in the Development of Microbial Double-Stranded RNA Production Systems for Application of RNA Interference in Agricultural Pest Control. Front. Bioeng. Biotechnol. 9:753790. doi: 10.3389/fbioe.2021.753790
Received: 05 August 2021; Accepted: 31 August 2021;
Published: 13 September 2021.
Edited by:
Tao Chen, Tianjin University, ChinaReviewed by:
Jie Shen, China Agricultural University, ChinaMing-Bo Wang, Commonwealth Scientific and Industrial Research Organisation (CSIRO), Australia
Copyright © 2021 Guan, Chu, Han, Miao and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xuexia Miao, xxm@cemps.ac.cn; Haichao Li, lihaichao@cemps.ac.cn
 Ruobing Guan
Ruobing Guan Dongdong Chu
Dongdong Chu Xinyi Han
Xinyi Han Xuexia Miao2*
Xuexia Miao2*  Haichao Li
Haichao Li