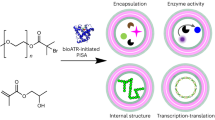
Abstract
Biological membrane-enclosed organelles are fascinating examples of spatially confined nanoreactors for biocatalytic transformations such as cascade reactions involving multiple enzymes; however, the fabrication of their synthetic mimics remains a considerable challenge. Here we demonstrate supramolecular chemistry-based bridging of two membranes leading to controlled fusion of exosomes that act as nanoreactors for effective biocatalytic cascades, with prolonged functionality inside of living cells. Exosome membrane proteins were chemically engineered with a catechol moiety to drive fusion by supramolecular complexation to bridge the membranes. This strategy successfully encapsulated multiple enzymes and assembled the minimal electron transport chain in the plasma membrane, leading to tuneable, enhanced catalytic cascade activity capable of ATP synthesis inside of tissue spheroids. This nanoreactor was functional for many hours after uptake into living cells, showed successful penetration into tissue spheroids and repaired the damaged region by supplying ATP, all of which represent an advance in the mimicking of nature’s own organelles.

This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
Data used to generate results in the current study are available from the corresponding author on reasonable request. Source data are provided with this paper.
References
Agapakis, C. M., Boyle, P. M. & Silver, P. A. Natural strategies for the spatial optimization of metabolism in synthetic biology. Nat. Chem. Biol. 8, 527–535 (2012).
Hammes, G. G. & Wu, C.-W. Regulation of enzyme activity. Science 172, 1205–1211 (1971).
Kondo, S. & Miura, T. Reaction-diffusion model as a framework for understanding biological pattern formation. Science 329, 1616–1620 (2010).
Lim, W. A. Designing customized cell signalling circuits. Nat. Rev. Mol. Cell Biol. 11, 393–403 (2010).
Han, D. et al. A cascade reaction network mimicking the basic functional steps of adaptive immune response. Nat. Chem. 7, 835–841 (2015).
Semenov, S. N. et al. Rational design of functional and tunable oscillating enzymatic networks. Nat. Chem. 7, 160–165 (2015).
Zhang, Y., Tsitkov, S. & Hess, H. Complex dynamics in a two-enzyme reaction network with substrate competition. Nat. Catal. 1, 276–281 (2018).
Chen, W.-H., Vázquez-González, M., Zoabi, A., Abu-Reziq, R. & Willner, I. Biocatalytic cascades driven by enzymes encapsulated in metal–organic framework nanoparticles. Nat. Catal. 1, 689–695 (2018).
Küchler, A., Yoshimoto, M., Luginbühl, S., Mavelli, F. & Walde, P. Enzymatic reactions in confined environments. Nat. Nanotechnol. 11, 409–420 (2016).
Vázquez-González, M., Wang, C. & Willner, I. Biocatalytic cascades operating on macromolecular scaffolds and in confined environments. Nat. Catal. 3, 256–273 (2020).
Einfalt, T. et al. Biomimetic artificial organelles with in vitro and in vivo activity triggered by reduction in microenvironment. Nat. Commun. 9, 1127 (2018).
Buddingh’, B. C. & van Hest, J. C. M. Artificial cells: synthetic compartments with life-like functionality and adaptivity. Acc. Chem. Res. 50, 769–777 (2017).
Staufer, O., Schröter, M., Platzman, I. & Spatz, J. P. Bottom-up assembly of functional intracellular synthetic organelles by droplet-based microfluidics. Small 16, 1906424 (2020).
van Oppen, L. M. P. E. et al. Biodegradable synthetic organelles demonstrate ROS shielding in human-complex-I-deficient fibroblasts. ACS Cent. Sci. 4, 917–928 (2018).
Reinkemeier, C. D., Girona, G. E. & Lemke, E. A. Designer membraneless organelles enable codon reassignment of selected mRNAs in eukaryotes. Science 363, eaaw2644 (2019).
Rideau, E., Dimova, R., Schwille, P., Wurm, F. R. & Landfester, K. Liposomes and polymersomes: a comparative review towards cell mimicking. Chem. Soc. Rev. 47, 8572–8610 (2018).
Parodi, A. et al. Synthetic nanoparticles functionalized with biomimetic leukocyte membranes possess cell-like functions. Nat. Nanotechnol. 8, 61–68 (2013).
Hu, C.-M. J. et al. Erythrocyte membrane-camouflaged polymeric nanoparticles as a biomimetic delivery platform. Proc. Natl Acad. Sci. USA 108, 10980 (2011).
Lee, K. Y. et al. Photosynthetic artificial organelles sustain and control ATP-dependent reactions in a protocellular system. Nat. Biotechnol. 36, 530–535 (2018).
Weiss, M. et al. Sequential bottom-up assembly of mechanically stabilized synthetic cells by microfluidics. Nat. Mater. 17, 89–96 (2018).
Yoo, J.-W., Irvine, D. J., Discher, D. E. & Mitragotri, S. Bio-inspired, bioengineered and biomimetic drug delivery carriers. Nat. Rev. Drug Discov. 10, 521–535 (2011).
Grzybowski, B. A. & Huck, W. T. S. The nanotechnology of life-inspired systems. Nat. Nanotechnol. 11, 585–592 (2016).
Miller, T. E. et al. Light-powered CO2 fixation in a chloroplast mimic with natural and synthetic parts. Science 368, 649–654 (2020).
Kumar, S. et al. Human platelet membrane functionalized microchips with plasmonic codes for cancer detection. Adv. Funct. Mater. 29, 1902669 (2019).
Kalluri, R. & LeBleu, V. S. The biology, function, and biomedical applications of exosomes. Science 367, eaau6977 (2020).
El Andaloussi, S., Mäger, I., Breakefield, X. O. & Wood, M. J. A. Extracellular vesicles: biology and emerging therapeutic opportunities. Nat. Rev. Drug Discov. 12, 347–357 (2013).
Armstrong, J. P. K., Holme, M. N. & Stevens, M. M. Re-engineering extracellular vesicles as smart nanoscale therapeutics. ACS Nano 11, 69–83 (2017).
Robbins, P. D. & Morelli, A. E. Regulation of immune responses by extracellular vesicles. Nat. Rev. Immunol. 14, 195–208 (2014).
Hindley, J. W. et al. Light-triggered enzymatic reactions in nested vesicle reactors. Nat. Commun. 9, 1093 (2018).
Ishmukhametov, R. R., Russell, A. N. & Berry, R. M. A modular platform for one-step assembly of multi-component membrane systems by fusion of charged proteoliposomes. Nat. Commun. 7, 13025 (2016).
Bolognesi, G. et al. Sculpting and fusing biomimetic vesicle networks using optical tweezers. Nat. Commun. 9, 1882 (2018).
Kumar, S., Michael, I. J., Park, J., Granick, S. & Cho, Y.-K. Cloaked exosomes: biocompatible, durable, and degradable encapsulation. Small 14, 1802052 (2018).
Stengel, G., Simonsson, L., Campbell, R. A. & Höök, F. Determinants for membrane fusion induced by cholesterol-modified DNA zippers. J. Phys. Chem. B 112, 8264–8274 (2008).
Jumeaux, C. et al. MicroRNA detection by DNA-mediated liposome fusion. ChemBioChem 19, 434–438 (2018).
Barlow, N., Chalmers, D. K., Williams-Noonan, B. J., Thompson, P. E. & Norton, R. S. Improving membrane permeation in the beyond rule-of-five space by using prodrugs to mask hydrogen bond donors. ACS Chem. Biol. 15, 2070–2078 (2020).
Stengel, G., Zahn, R. & Höök, F. DNA-induced programmable fusion of phospholipid vesicles. J. Am. Chem. Soc. 129, 9584–9585 (2007).
Collins, D. J., Neild, A., deMello, A., Liu, A.-Q. & Ai, Y. The Poisson distribution and beyond: methods for microfluidic droplet production and single cell encapsulation. Lab Chip 15, 3439–3459 (2015).
Huang, F. et al. Near-infrared light-activated membrane fusion for cancer cell therapeutic applications. Chem. Sci. 11, 5592–5600 (2020).
Piffoux, M., Silva, A. K. A., Wilhelm, C., Gazeau, F. & Tareste, D. Modification of extracellular vesicles by fusion with liposomes for the design of personalized biogenic drug delivery systems. ACS Nano 12, 6830–6842 (2018).
Leidal, A. M. et al. The LC3-conjugation machinery specifies the loading of RNA-binding proteins into extracellular vesicles. Nat. Cell Biol. 22, 187–199 (2020).
Fuentes, P. et al. ITGB3-mediated uptake of small extracellular vesicles facilitates intercellular communication in breast cancer cells. Nat. Commun. 11, 4261 (2020).
Joshi, B. S., de Beer, M. A., Giepmans, B. N. G. & Zuhorn, I. S. Endocytosis of extracellular vesicles and release of their cargo from endosomes. ACS Nano 14, 4444–4455 (2020).
Xu, Y. et al. Nanozyme-catalyzed cascade reactions for mitochondria-mimicking oxidative phosphorylation. Angew. Chem. Int. Ed. 58, 5572–5576 (2019).
Jeong, S., Nguyen, H. T., Kim, C. H., Ly, M. N. & Shin, K. Toward artificial cells: novel advances in energy conversion and cellular motility. Adv. Funct. Mater. 30, 1907182 (2020).
Schütt, F., Aretz, S., Auffarth, G. U. & Kopitz, J. Moderately reduced ATP levels promote oxidative stress and debilitate autophagic and phagocytic capacities in human RPE cells. Investig. Ophthalmol. Vis. Sci. 53, 5354–5361 (2012).
Lenzini, S., Bargi, R., Chung, G. & Shin, J.-W. Matrix mechanics and water permeation regulate extracellular vesicle transport. Nat. Nanotechnol. 15, 217–223 (2020).
Yong, T. et al. Tumor exosome-based nanoparticles are efficient drug carriers for chemotherapy. Nat. Commun. 10, 3838 (2019).
Borkowska, M. et al. Targeted crystallization of mixed-charge nanoparticles in lysosomes induces selective death of cancer cells. Nat. Nanotechnol. 15, 331–341 (2020).
Feder, T. J., Brust-Mascher, I., Slattery, J. P., Baird, B. & Webb, W. W. Constrained diffusion or immobile fraction on cell surfaces: a new interpretation. Biophys. J. 70, 2767–2773 (1996).
Nandi, A., Heinrich, D. & Lindner, B. Distributions of diffusion measures from a local mean-square displacement analysis. Phys. Rev. E 86, 021926 (2012).
Ishmukhametov, R. R., Galkin, M. A. & Vik, S. B. Ultrafast purification and reconstitution of His-tagged cysteine-less Escherichia coli F1Fo ATP synthase. Biochim. Biophys. Acta 1706, 110–116 (2005).
Rumbley, J. N., Furlong Nickels, E. & Gennis, R. B. One-step purification of histidine-tagged cytochrome bo3 from Escherichia coli and demonstration that associated quinone is not required for the structural integrity of the oxidase. Biochim. Biophys. Acta 1340, 131–142 (1997).
Acknowledgements
This work was supported by the taxpayers of South Korea through the Institute for Basic Science (IBS-R020-D1).
Author information
Authors and Affiliations
Contributions
S.K., M.K., I.J.M. and Y.-K.C. conceived the research. S.K. designed the approach and characterized all the materials. S.K., M.K. and Y.-K.C. prepared the manuscript. I.J.M. performed the majority of microfluidic device experiments. Y.C. and J.Y.K. performed the exosome tracking experiments and analysed the data. I.K. performed exosome characterization. At all the stages, Y.-K.C. supervised the research, designed experiments, and wrote the paper. All authors read and corrected the manuscript.
Corresponding author
Ethics declarations
Competing interests
I.B.S. has filed a patent (KR 10-2021-0072646) based on this work and S.K. and Y.K.C. are named as inventors. UNIST has filed patents on ExoDisc and Y.K.C. is named as an inventor, which are licensed to LabSpinner. The other authors declare no competing interests.
Additional information
Peer review information Nature Catalysis thanks Gregor Fuhrmann, Mei He and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
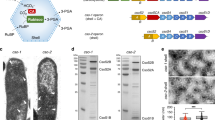
Extended Data Fig. 1 Characterization of exosomes and CEx.
(a) Characterization of the size of exosomes, CEx, and FEx using nanoparticles tracking analysis (NTA) for 5 independent experiments. Data represent mean ± SD and two-tailed unpaired Student’s t-test was used for comparing two groups (b) Characterization of the size distribution of exosomes from transmission electron microscopy (TEM) images. The histogram reports the size (nm) of exosomes vs frequency (%). For exosomes, 10 data points from n = 64 different TEM images; for CEx, 10 data points from n = 55 different TEM images; and for FEx, 10 data points from n = 56 different TEM images. (c) Zeta-potential of exosomes and CEx show that the average value is -18.5 ± 3.2 and -23.7 ± 2.9 eV, respectively (n = 5 independent experiments). After catechol modification, the zeta-potential distribution of CEx shows a negative shift compared to that of exosomes. Data represent mean ± SD and two-tailed unpaired Student’s t-test was used for comparing two groups (d) Characterization of the abundant exosomal proteins in each successive step during formation of fused exosomes (FEx) nanoreactor, analyzed by Western blot. Tetraspanin protein markers such as CD9, CD63, and CD81 are often used to characterize exosomes. From the results of Western blot analysis, it shows the presence of all the exosomes marker in control (exosomes), catechol integrated exosomes (CEx-1, CEx-2), and FEx, n = 3 independent experiments.
Extended Data Fig. 2 Comparison of the catalytic cascade after purification of FEx nanoreactor from free enzymes.
(a) Representative fluorescence spectra of resorufin generated by the two-enzyme cascade, under aerobic conditions, in the presence of FEx-GOx-HRP (black line) and treated with pronase (red line). (b) A photograph of Exodisc with 6 identical units for analysis of the CEx samples. Image on the right shows the detailed microfluidic features of the device including a sample loading chamber, filtration unit, and waste chamber. (c) A photograph of the table top-sized spinning device. (d) A schematic for the cross-sectional view of the filter unit of Exodisc. (e) Representative fluorescence intensity of resorufin generated by the two-enzyme cascade in the presence of CEx-GOx, CEx-HRP, CEx-GOx-HRP, and FEx-GOx-HRP purified from ultracentrifugation (left) and Exodisc (right). Data represent mean ± SD; n = 3 independent fusion experiments; two-tailed unpaired Student’s t-test was used for comparing two groups.
Extended Data Fig. 3 Co-localization of FEx nanoreactors with endosomes and lysosomes after cellular uptake.
(a) Representative time-lapse high resolution confocal fluorescence microscopy images of MCF-10A cells after treatment with FEx (green), and counter-stained for endosomes (pHrodo red dextran). Scale bar, 10 µm. n = 7 independent experiments. (b) Plot of Pearson correlation coefficient (PCCEndosome-FEx) values over time for red (Endosome) and green (FEx) fluorescence (at 10 min, 20 min, 30 min, 40 min, 50 min, and 60 min). Data represent mean ± SD of n = 7 independent experiments. (c) Representative time-lapse high resolution confocal fluorescence microscopy images of MCF-10A cells after treatment with FEx (green), fixed, and counter-stained with LysotrackerTM deep red (magenta) to study the cellular uptake mechanism. Scale bar, 10 µm. n = 7 independent experiments. (d) Plot of Pearson correlation coefficient (PCCLysosome-FEx) values over time for magenta (lysosome) and green (FEx) fluorescence (at 10 min, 20 min, 30 min, 40 min, 50 min, and 60 min). Data represent mean ± SD of n = 10 independent experiments.
Extended Data Fig. 4 Co-localization of FEx nanoreactor with endosomes after cellular uptake.
(a) Representative time-lapse high-resolution confocal fluorescence microscopy images of MCF-10A cells after treatment with FEx (green), fixed, and counter-stained for early endosomal antigen 1 marker EEA1 (red). Scale bar, 10 µm. n = 10 independent experiments. (b) Plot of Pearson correlation coefficient (PCCEndosome-FEx) values over time for red (Endosome) and green (FEx) fluorescence (at 10 min, 1 h, 2 h, 4 h, 8 h, and 12 h). Data represent mean ± SD; n = 10 independent experiments. (c) Representative time-lapse western blots of endosomal fractions for cellular uptake of FEx nanoreactor. The figure shows the relative amount of the CD63 proteins for FEx nanoreactor (53 kDa) and EEA1 protein for endosomes (162 kDa) after endosomal isolation from MCF-10A cells using MinuteTM Endosome Isolation kit. n = 3 independent experiments.
Extended Data Fig. 5 Co-localization of FEx nanoreactor with lysosomes after cellular uptake.
(a) Representative time-lapse high-resolution confocal fluorescence microscopy images of MCF-10A cells after treatment with FEx (green), fixed, and counter-stained for lysosome-associated membrane glycoprotein 1 (magenta) to study the intracellular mechanism. Scale bar, 10 µm. n = 10 independent experiments. (b) Plot of Pearson correlation coefficient (PCCLysosome-FEx) values over time for magenta (lysosome) and green (FEx) fluorescence (at 10 min, 1 h, 2 h, 4 h, 8 h, and 12 h). Data represent mean ± SD of n = 10 independent experiments. (c) Representative time-lapse western blots of lysosomal fractions for cellular uptake of FEx nanoreactor. The figure shows the relative amount of the LAMP1 proteins and Rab7 protein for lysosomes (110 kDa, 23 kDa respectively) and CD63 (53 kDa) for FEx nanoreactors from MCF-10A cells. Lysosomal fraction was isolated using MinuteTM Lysosome Isolation kit, n = 3 independent experiments.
Extended Data Fig. 6 Stability and integrity of FEx-GOx-HRP nanoreactor after cellular integration.
(a) Representative confocal 3D fluorescence microscopy images for co-localization study of the FEx-GOx-HRP (encapsulated FITC-labelled GOx at 488 nm and rhodamine B-labelled HRP at 561 nm) nanoreactor upon cellular uptake. Confocal fluorescence microscopic analysis after incubating FEx with MCF-10A cells for 24 h. FITC-GOx and Rhodamine-HRP overlay images. n = 3 independent experiments. (b) Time-course study of Pearson’s correlation coefficient (PCC). To study the integrity of the content (FITC-labelled GOx and rhodamine B-labelled HRP) inside FEx nanoreactor. Data represent the time-lapse value (at 0-24 h) of PCCFITC-Rhodamine is computed. Data represent mean ± SD and are based on three independent experiments (n = 47 MCF-10A cells). (c) Motility of FEx-GOx-HRP nanoreactor after cellular integration. Representative Live-cell tracking confocal fluorescence microscopy images of FEx-GOx-HRP (FITC-labelled GOx) after cellular uptake by the human breast epithelial cells MCF-10A after 48 h where FEx movement was indicated by arrowhead. n = 2 independent experiments. (d) Representative confocal fluorescence images (bright field, red, green, magenta, and overlay) of the MCF-10A cells after 48 h of incubation with FEx and immunofluorescent staining of EEA1 (red) and LAMP1 (magenta). Scale bar, 10 µm. Co-localization of FEx (green) with Endosomes (red) and Lysosomes (magenta) after 48 h. Panels 1-6 show high-resolution confocal fluorescence images of sections indicated on the overlay image. Scale bar, 2 µm. n = 3 independent experiments. (e) Representative transmission electron micrographs (TEM) of MCF-10A cells after incubation with immunogold-labelled FEx nanoreactor (indicated by arrowhead) for 48 h, confirming the presence of intact FEx nanoreactor. Scale bar, 5 µm. Extended TEM images scale bar, 500 nm. n = 2 independent experiments.
Extended Data Fig. 7 Bioorthogonal catalysis in fused exosomes.
(a) Catalytic conversion of proRho to free rhodamine inside the cells by using the FEx-Pd-proRho nanoreactor. The FEx-Pd-proRho nanoreactor was prepared by fusing palladium nanoparticle- (Pd NPs) loaded exosomes (CEx-Pd) and bis-allyloxycarbonyl-protected rhodamine- (proRho) loaded exosomes (CEx-proRho). (b) Representative TEM images of FEx-Pd-proRho. Inset shows magnified image of selection showing the presence of Pd nanostructures of size ~6 nm. Scale bar, 10 nm. n = 2 independent experiments. (c) Fluorescence emission spectra (λEx = 498 nm) for demasking of Rhodamine in FEx and CEx. (d) Change in fluorescence intensity (%) (at 527 nm) of demasking reaction as the function of time in the FEx-Pd-proRho and CEx-proRho. (e) Representative 3D Confocal fluorescence images with FITC filter (representing rhodamine fluorescence), DAPI (nuclei staining) and merged images after cellular uptake of FEx-Pd-proRho and CEx-proRho. n = 3 independent experiments. (f) Flow cytometry analysis for monitoring the demasking reaction inside the MCF-10A cells with FEx. (g) Catalytic conversion of pro-drug to drug with the FEx-Pd-proFU nanoreactor. Schematic for the activation of anticancer drug 5-fluorouracil (pro-FU) using FEx-Pd- proFU nanoreactor. This drug converts cytotoxic nucleotide-metabolites via functionalization on its N1-position followed by inhibition of thymidylate synthase and incorporation into RNA- and DNA-strands to disrupt cellular function (h) The cell viability of MCF-10A cells after incubating them with different concentrations of the nanoreactors FEx-Pd- proFU and CEx-proFU (10 µg mL-1, 30 µg mL-1, 60 µg mL-1, and 90 µg mL-1). Data represent mean ± SD; n = 3 independent experiments; two-tailed unpaired Student’s t-test was used for comparing two groups.
Extended Data Fig. 8 Estimation of ATP.
(a) Estimation of ATP in FEx-1 nanoreactors by the luciferin-luciferase assay. A standard calibration curve was obtained by monitoring luminescence intensity (luciferin to oxy-luciferin) by varying the amount of ATP (0 to 1000 nM). (b) Time-dependent changes in luminescence intensity were measured using the luciferin-luciferase assay by the treatment of FEx nanoreactors with DTT, glucose, or DTT-glucose. After the assembly of ATP synthase and bo3 oxidase into the membranes of the FEx nanoreactors as energy modules, the electron transport chain was triggered by the addition of dithiothreitol (DTTred), which reduced coenzyme Q1 (Q1H) to make it available to bo3 oxidase and encapsulated GOx/HRP inside FEx-1 nanoreactors by addition of glucose. (c) Estimation of ROS production during the functioning of FEx-1 nanoreactors inside living cells by using the ROS assay kit. A standard calibration curve was obtained by monitoring the fluorescence intensity of dichlorodihydrofluorescein diacetate (excitation at 485 nm and emission at 535 nm) after treatment with varying concentrations of tert-butyl hydrogen peroxide (0 to 50 µM). These results demonstrate that the increase in ROS production was proportional to the increase in fluorescence intensity. (d) MCF-10A cells were treated with different concentrations of FEx-1 nanoreactors (10, 50, and 80 µg/mL), and the fluorescence intensity of dichlorodihydrofluorescein diacetate was measured to calculate ROS levels. (e) Schematic of controls for direct loading of the enzymes into exosomes. ATP production using the luciferin luciferase assay of different condition controls; control (only exosome), (A), (B), and FEx-1 after 3 h. In the first control (A), GOx, HRP, ATPsynthase, and bo3Oxidase were used all together to encapsulated inside the exosome. Their amount was quantified by BCA Protein Assay kit after removal of free enzymes by ultracentrifugation. In the second control (B), all the enzymes (GOx, HRP, ATPsynthase and bo3Oxidase) were encapsulated one by one after the other with each time washing between steps, respectively. (f) Luminescence intensity of multi-enzyme cascade reaction to produce the bioenergy in only exosomes, (A), and (B). ATP production was measured by luciferin-luciferase assay. Data represent mean ± SD of n = 3 independent experiments; two-tailed unpaired Student’s t-test was used for comparing two groups.
Supplementary information
Supplementary Information
Supplementary Methods and Figs. 1–8.
41929_2021_669_MOESM3_ESM.mp4
Supplementary Video 1 Microfluidic chip used for droplet generation. Droplet-based microfluidics device was used to perform high-throughput generation of the monodisperse cell-sized droplets (~10 µm) where one aqueous phase contained the CEx solutions and the other phase contained FeCl3·6H2O that met at the flow-focusing junction and were delivered into the oil stream for breaking into uniform droplets.
41929_2021_669_MOESM4_ESM.mp4
Supplementary Video 2 CEx transportation dynamics. Tracking data overlaid with imaging data for representation of transport of CEx inside the cells. The video confirms the CEx nanoreactor transportation dynamics inside the living cells.
41929_2021_669_MOESM5_ESM.mp4
Supplementary Video 3 FEx transportation dynamics. Tracking data overlaid with imaging data for representation of transport of FEx inside the cells. The video confirms the FEx nanoreactor transportation dynamics inside the living cells.
Source data
Source Data Extended Data Fig. 1
Full western blots for Extended Data Fig. 1d.
Source Data Extended Data Fig. 4
Full western blots for Extended Data Fig. 4c.
Source Data Extended Data Fig. 5
Full western blots for Extended Data Fig. 5c.
Source Data Extended Data Fig. 7
FACS data for Extended Data Fig. 7f.
Rights and permissions
About this article
Cite this article
Kumar, S., Karmacharya, M., Michael, I.J. et al. Programmed exosome fusion for energy generation in living cells. Nat Catal 4, 763–774 (2021). https://doi.org/10.1038/s41929-021-00669-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41929-021-00669-z
This article is cited by
-
Gastric cancer and mesenchymal stem cell-derived exosomes: from pro-tumorigenic effects to anti-cancer vehicles
Archives of Pharmacal Research (2024)
-
Recent advances in extracellular vesicle engineering and its applications to regenerative medicine
Biomaterials Research (2023)