Advanced Molecular Dynamics Approaches to Model a Tertiary Complex APRIL/TACI with Long Glycosaminoglycans
Abstract
:1. Introduction
2. Materials and Methods
2.1. Structures
2.2. Molecular Docking
2.3. Molecular Dynamics
2.4. Binding Free Energy Calculations
3. Results and Discussion
3.1. Docking Heparin dp24 to APRIL–BCMA and APRIL–TACI Complexes Using Autodock3
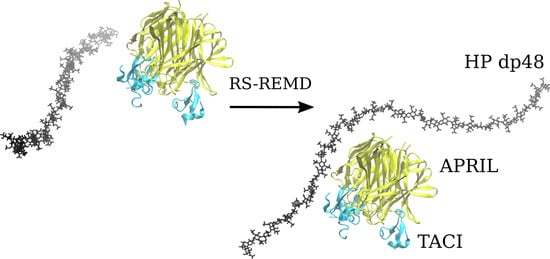
3.2. Docking Heparin dp24 and dp48 to APRIL–BCMA and APRIL–TACI Complexes Using RS-REMD
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Paiardi, G.; Milanesi, M.; Wade, R.C.; D’ursi, P.; Rusnati, M. A Bittersweet Computational Journey among Glycosaminoglycans. Biomolecules 2021, 11, 739. [Google Scholar] [CrossRef]
- Habuchi, H.; Habuchi, O.; Kimata, K. Sulfation Pattern in Glycosaminoglycan: Does It Have a Code? Glycoconj. J. 2004, 21, 47–52. [Google Scholar] [CrossRef]
- Varki, A.; Cummings, R.D.; Esko, J.D.; Stanley, P.; Hart, G.W.; Aebi, A.; Darvill, A.G.; Kinoshita, T.; Packer, N.H.; Prestegard, J.H.; et al. Essentials of Glycobiology, 3rd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2015. [Google Scholar]
- Bu, C.; Jin, L. NMR Characterization of the Interactions between Glycosaminoglycans and Proteins. Front. Mol. Biosci. 2021, 8, 165. [Google Scholar] [CrossRef]
- Clerc, O.; Mariethoz, J.; Rivet, A.; Lisacek, F.; Pérez, S.; Ricard-Blum, S. A Pipeline to Translate Glycosaminoglycan Sequences into 3D Models. Application to the Exploration of Glycosaminoglycan Conformational Space. Glycobiology 2019, 29, 36–44. [Google Scholar] [CrossRef]
- Vallet, S.D.; Clerc, O.; Ricard-Blum, S. Glycosaminoglycan–Protein Interactions: The First Draft of the Glycosaminoglycan Interactome. J. Histochem. Cytochem. 2020, 69, 93–104. [Google Scholar] [CrossRef] [PubMed]
- Van Der Smissen, A.; Samsonov, S.; Hintze, V.; Scharnweber, D.; Moeller, S.; Schnabelrauch, M.; Pisabarro, M.T.; Anderegg, U. Artificial Extracellular Matrix Composed of Collagen i and Highly Sulfated Hyaluronan Interferes with TGFβ1 Signaling and Prevents TGFβ1-Induced Myofibroblast Differentiation. Acta Biomater. 2013, 9, 7775–7786. [Google Scholar] [CrossRef]
- Pichert, A.; Samsonov, S.A.; Theisgen, S.; Thomas, L.; Baumann, L.; Schiller, J.; Beck-Sickinger, A.G.; Huster, D.; Pisabarro, M.T. Characterization of the Interaction of Interleukin-8 with Hyaluronan, Chondroitin Sulfate, Dermatan Sulfate and Their Sulfated Derivatives by Spectroscopy and Molecular Modeling. Glycobiology 2012, 22, 134–145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bojarski, K.K.; Samsonov, S.A. Role of Oligosaccharide Chain Polarity in Protein-Glycosaminoglycan Interactions. J. Chem. Inf. Model. 2021, 61, 455–466. [Google Scholar] [CrossRef]
- Gallagher, J. Fell-Muir Lecture: Heparan Sulphate and the Art of Cell Regulation: A Polymer Chain Conducts the Protein Orchestra. Int. J. Exp. Pathol. 2015, 96, 203–231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kjellén, L.; Lindahl, U. Specificity of Glycosaminoglycan–Protein Interactions. Curr. Opin. Struct. Biol. 2018, 50, 101–108. [Google Scholar] [CrossRef]
- Imberty, A.; Lortat-Jacob, H.; Pérez, S. Structural View of Glycosaminoglycan–Protein Interactions. Carbohydr. Res. 2007, 342, 430–439. [Google Scholar] [CrossRef]
- Marcisz, M.; Huard, B.; Lipska, A.G.; Samsonov, S.A. Further Analyses of APRIL/APRIL-Receptor/Glycosaminoglycan Interactions by Biochemical Assays Linked to Computational Studies. Glycobiology 2021, 31, 772–786. [Google Scholar] [CrossRef]
- Uciechowska-Kaczmarzyk, U.; Babik, S.; Zsila, F.; Bojarski, K.K.; Beke-Somfai, T.; Samsonov, S.A. Molecular Dynamics-Based Model of {VEGF}-{A} and Its Heparin Interactions. J. Mol. Graph. Model. 2018, 82, 157–166. [Google Scholar] [CrossRef]
- Gandhi, N.S.; Mancera, R.L. Molecular Dynamics Simulations of CXCL-8 and Its Interactions with a Receptor Peptide, Heparin Fragments, and Sulfated Linked Cyclitols. J. Chem. Inf. Model. 2011, 51, 335–358. [Google Scholar] [CrossRef]
- Joseph, P.R.B.; Mosier, P.D.; Desai, U.R.; Rajarathnam, K. Solution NMR Characterization of Chemokine CXCL8/IL-8 Monomer and Dimer Binding to Glycosaminoglycans: Structural Plasticity Mediates Differential Binding Interactions. Biochem. J. 2015, 472, 121–133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clausen, T.M.; Sandoval, D.R.; Spliid, C.B.; Pihl, J.; Perrett, H.R.; Painter, C.D.; Narayanan, A.; Majowicz, S.A.; Kwong, E.M.; McVicar, R.N.; et al. SARS-CoV-2 Infection Depends on Cellular Heparan Sulfate and ACE2. Cell 2020, 183, 1043–1057.e15. [Google Scholar] [CrossRef] [PubMed]
- Schuurs, Z.P.; Hammond, E.; Elli, S.; Rudd, T.R.; Mycroft-West, C.J.; Lima, M.A.; Skidmore, M.A.; Karlsson, R.; Chen, Y.H.; Bagdonaite, I.; et al. Evidence of a Putative Glycosaminoglycan Binding Site on the Glycosylated SARS-CoV-2 Spike Protein N-Terminal Domain. Comput. Struct. Biotechnol. J. 2021, 19, 2806–2818. [Google Scholar] [CrossRef] [PubMed]
- Uciechowska-Kaczmarzyk, U.; de Beauchene, I.; Samsonov, S.A. Docking Software Performance in Protein-Glycosaminoglycan Systems. J. Mol. Graph. Model. 2019, 90, 42–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samsonov, S.A.; Gehrcke, J.P.; Pisabarro, M.T. Flexibility and Explicit Solvent in Molecular-Dynamics-Based Docking of Protein-Glycosaminoglycan Systems. J. Chem. Inf. Model. 2014, 54, 582–592. [Google Scholar] [CrossRef]
- Samsonov, S.A.; Zacharias, M.; Chauvot de Beauchene, I. Modeling Large Protein–Glycosaminoglycan Complexes Using a Fragment-Based Approach. J. Comput. Chem. 2019, 40, 1429–1439. [Google Scholar] [CrossRef] [Green Version]
- Siebenmorgen, T.; Engelhard, M.; Zacharias, M. Prediction of Protein–Protein Complexes Using Replica Exchange with Repulsive Scaling. J. Comput. Chem. 2020, 41, 1436–1447. [Google Scholar] [CrossRef] [Green Version]
- Siebenmorgen, T.; Zacharias, M. Efficient Refinement and Free Energy Scoring of Predicted Protein-Protein Complexes Using Replica Exchange with Repulsive Scaling. J. Chem. Inf. Model. 2020, 60, 5552–5562. [Google Scholar] [CrossRef] [PubMed]
- Maszota-Zieleniak, M.; Marcisz, M.; Kogut, M.M.; Siebenmorgen, T.; Zacharias, M.; Samsonov, S.A. Evaluation of Replica Exchange with Repulsive Scaling Approach for Docking Glycosaminoglycans. J. Comput. Chem. 2021, 42, 1040–1453. [Google Scholar] [CrossRef] [PubMed]
- Hahne, M.; Kataoka, T.; Schröter, M.; Hofmann, K.; Irmler, M.; Bodmer, J.L.; Schneider, P.; Bornand, T.; Holler, N.; French, L.E.; et al. APRIL, a New Ligand of the Tumor Necrosis Factor Family, Stimulates Tumor Cell Growth. J. Exp. Med. 1998, 188, 1185–1890. [Google Scholar] [CrossRef] [Green Version]
- Hendriks, J.; Planelles, L.; de Jong-Odding, J.; Hardenberg, G.; Pals, S.T.; Hahne, M.; Spaargaren, M.; Medema, J.P. Heparan Sulfate Proteoglycan Binding Promotes APRIL-Induced Tumor Cell Proliferation. Cell Death Differ. 2005, 12, 637–648. [Google Scholar] [CrossRef] [Green Version]
- Ingold, K.; Zumsteg, A.; Tardivel, A.; Huard, B.; Steiner, Q.G.; Cachero, T.G.; Qiang, F.; Gorelik, L.; Kalled, S.L.; Acha-Orbea, H.; et al. Identification of Proteoglycans as the APRIL-Specific Binding Partners. J. Exp. Med. 2005, 201, 1375–1383. [Google Scholar] [CrossRef] [Green Version]
- Baert, L.; Benkhoucha, M.; Popa, N.; Ahmed, M.C.; Manfroi, B.; Boutonnat, J.; Sturm, N.; Raguenez, G.; Tessier, M.; Casez, O.; et al. A Proliferation-Inducing Ligand–Mediated Anti-Inflammatory Response of Astrocytes in Multiple Sclerosis. Ann. Neurol. 2019, 85, 406–420. [Google Scholar] [CrossRef]
- Kimberley, F.C.; Medema, J.P.; Hahne, M. APRIL in B-Cell Malignancies and Autoimmunity. Results Probl. Cell Differ. 2009, 49, 161–182. [Google Scholar] [CrossRef]
- Kimberley, F.C.; Van Bostelen, L.; Cameron, K.; Hardenberg, G.; Marquart, J.A.; Hahne, M.; Medema, J.P. The Proteoglycan (Heparan Sulfate Proteoglycan) Binding Domain of APRIL Serves as a Platform for Ligand Multimerization Andcross-Linking. FASEB J. 2009, 23, 1584–1595. [Google Scholar] [CrossRef] [PubMed]
- Huard, B.; McKee, T.; Bosshard, C.; Durual, S.; Matthes, T.; Myit, S.; Donze, O.; Frossard, C.; Chizzolini, C.; Favre, C.; et al. APRIL Secreted by Neutrophils Binds to Heparan Sulfate Proteoglycans to Create Plasma Cell Niches in Human Mucosa. J. Clin. Investig. 2008, 118, 2887–2895. [Google Scholar] [CrossRef] [PubMed]
- Bischof, D.; Elsawa, S.F.; Mantchev, G.; Yoon, J.; Michels, G.E.; Nilson, A.; Sutor, S.L.; Platt, J.L.; Ansell, S.M.; Von Bulow, G.; et al. Selective Activation of TACI by Syndecan-2. Blood 2006, 107, 3235–3242. [Google Scholar] [CrossRef]
- Moreaux, J.; Sprynski, A.C.; Dillon, S.R.; Mahtouk, K.; Jourdan, M.; Ythier, A.; Moine, P.; Robert, N.; Jourdan, E.; Rossi, J.F.; et al. APRIL and TACI Interact with Syndecan-1 on the Surface of Multiple Myeloma Cells to form an Essential Survival Loop. Eur. J. Haematol. 2009, 83, 119–129. [Google Scholar] [CrossRef]
- Sakurai, D.; Hase, H.; Kanno, Y.; Kojima, H.; Okumura, K.; Kobata, T. TACI Regulates IgA Production by APRIL in Collaboration with HSPG. Blood 2007, 109, 2961–2967. [Google Scholar] [CrossRef] [Green Version]
- Kowalczyk-Quintas, C.; Willen, D.; Willen, L.; Golob, M.; Schuepbach-Mallepell, S.; Peter, B.; Eslami, M.; Vigolo, M.; Broly, H.; Samy, E.; et al. No Interactions between Heparin and Atacicept, an Antagonist of B Cell Survival Cytokines. Br. J. Pharmacol. 2019, 176, 4019–4033. [Google Scholar] [CrossRef] [Green Version]
- Hymowitz, S.G.; Patel, D.R.; Wallweber, H.J.A.; Runyon, S.; Yan, M.; Yin, J.P.; Shriver, S.K.; Gordon, N.C.; Pan, B.; Skelton, N.J.; et al. Structures of APRIL-Receptor Complexes: Like BCMA, TACI Employs Only a Single Cysteine-Rich Domain for High Affinity Ligand Binding. J. Biol. Chem. 2005, 280, 7218–7227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pichert, A.; Schlorke, D.; Franz, S.; Arnhold, J. Functional Aspects of the Interaction between Interleukin-8 and Sulfated Glycosaminoglycans. Biomatter 2012, 2, 142–148. [Google Scholar] [CrossRef] [Green Version]
- Kirschner, K.N.; Yongye, A.B.; Tschampel, S.M.; González-Outeiriño, J.; Daniels, C.R.; Foley, B.L.; Woods, R.J. GLYCAM06: A Generalizable Biomolecular Force Field. Carbohydrates. J. Comput. Chem. 2008, 29, 622–655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huige, C.J.M.; Altona, C. Force Field Parameters for Sulfates and Sulfamates Based on Ab Initio Calculations: Extensions of AMBER and CHARMm Fields. J. Comput. Chem. 1995, 16, 56–79. [Google Scholar] [CrossRef]
- Samsonov, S.A.; Pisabarro, M.T. Computational Analysis of Interactions in Structurally Available Protein–Glycosaminoglycan Complexes. Glycobiology 2016, 26, 850–861. [Google Scholar] [CrossRef] [Green Version]
- Ester, M.; Kriegel, H.-P.; Sander, J.; Xu, X. A Density-Based Algorithm for Discovering Clustersin Large Spatial Databases with Noise. AAAI 1996, 96, 226–231. [Google Scholar]
- Maier, J.A.; Martinez, C.; Kasavajhala, K.; Wickstrom, L.; Hauser, K.E.; Simmerling, C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 2015, 16, 528–552. [Google Scholar] [CrossRef] [Green Version]
- Case, D.A.; Ben-Shalom, I.Y.; Brozell, S.R.; Cerutti, D.S.; Cheatham, T.E., III; Cruzeiro, V.W.D.; Darden, T.A.; Duke, R.E.; Ghoreishi, D.; Gilson, M.K.; et al. AMBER16; University of California: San Francisco, CA, USA, 2018. [Google Scholar]
- Darden, T.; York, D.; Pedersen, L. Particle mesh Ewald: An N⋅log(N) method for Ewald sums in large systems. J. Phys. Chem. 1993, 98, 10089. [Google Scholar] [CrossRef] [Green Version]
- Ryckaert, J.-P.; Ciccotti, G.; Berendsen, J.C. Numerical integration of the cartesian equations of motion of a system with constraints: Molecular dynamics of n-alkanes. J. Comp. Phys. 1977, 23, 327–341. [Google Scholar] [CrossRef] [Green Version]
- Onufriev, A.; Case, D.A.; Bashford, D. Effective Born Radii in the Generalized Born Approximation: The Importance of Being Perfect. J. Comput. Chem. 2002, 23, 1297–1304. [Google Scholar] [CrossRef] [PubMed]
- Homeyer, N.; Gohlke, H. Free Energy Calculations by the Molecular Mechanics Poisson−Boltzmann Surface Area Method. Mol. Inform. 2012, 31, 114–122. [Google Scholar] [CrossRef] [PubMed]
- Gadhhi, N.S.; Mancera, R.L. Free energy calculations of glycosaminoglycan–protein interactions. Glycobiology 2009, 19, 1103–1115. [Google Scholar] [CrossRef] [Green Version]
- Nivedha, A.K.; Makeneni, S.; Foley, B.L.; Tessier, M.B.; Woods, R.J. Importance of Ligand Conformational Energies in Carbohydrate Docking: Sorting the Wheat from the Chaff. J. Comput. Chem. 2014, 35, 526–539. [Google Scholar] [CrossRef] [Green Version]





Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marcisz, M.; Maszota-Zieleniak, M.; Huard, B.; Samsonov, S.A. Advanced Molecular Dynamics Approaches to Model a Tertiary Complex APRIL/TACI with Long Glycosaminoglycans. Biomolecules 2021, 11, 1349. https://doi.org/10.3390/biom11091349
Marcisz M, Maszota-Zieleniak M, Huard B, Samsonov SA. Advanced Molecular Dynamics Approaches to Model a Tertiary Complex APRIL/TACI with Long Glycosaminoglycans. Biomolecules. 2021; 11(9):1349. https://doi.org/10.3390/biom11091349
Chicago/Turabian StyleMarcisz, Mateusz, Martyna Maszota-Zieleniak, Bertrand Huard, and Sergey A. Samsonov. 2021. "Advanced Molecular Dynamics Approaches to Model a Tertiary Complex APRIL/TACI with Long Glycosaminoglycans" Biomolecules 11, no. 9: 1349. https://doi.org/10.3390/biom11091349