Abstract
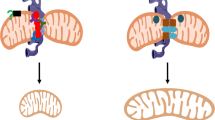
Mitochondria are the main site for generating reactive oxygen species, which are key players in diverse biological processes. However, the molecular pathways of redox signal transduction from the matrix to the cytosol are poorly defined. Here we report an inside-out redox signal of mitochondria. Cysteine oxidation of MIC60, an inner mitochondrial membrane protein, triggers the formation of disulfide bonds and the physical association of MIC60 with Miro, an outer mitochondrial membrane protein. The oxidative structural change of this membrane-crossing complex ultimately elicits cellular responses that delay mitophagy, impair cellular respiration and cause oxidative stress. Blocking the MIC60–Miro interaction or reducing either protein, genetically or pharmacologically, extends lifespan and health-span of healthy fruit flies, and benefits multiple models of Parkinson’s disease and Friedreich’s ataxia. Our discovery provides a molecular basis for common treatment strategies against oxidative stress.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
Data availability
Further information and reagents are available from the corresponding author upon reasonable request. Source data are provided with this paper. The Uniprot Drosophila melanogaster database was used.
References
Scialo, F. et al. Mitochondrial ROS produced via reverse electron transport extend animal lifespan. Cell Metab. 23, 725–734 (2016).
Xiao, H. et al. A quantitative tissue-specific landscape of protein redox regulation during aging. Cell 180, 968–983 (2020).
Reczek, C. R. & Chandel, N. S. ROS-dependent signal transduction. Curr. Opin. Cell Biol. 33, 8–13 (2015).
Sakowska, P. et al. The oxidation status of Mic19 regulates MICOS assembly. Mol. Cell. Biol. 35, 4222–4237 (2015).
Alvarez-Zaldiernas, C. et al. Cellular redox systems impact the aggregation of Cu,Zn superoxide dismutase linked to familial amyotrophic lateral sclerosis. J. Biol. Chem. 291, 17197–17208 (2016).
Pfanner, N., Warscheid, B. & Wiedemann, N. Mitochondrial proteins: from biogenesis to functional networks. Nat. Rev. Mol. Cell Biol. 20, 267–284 (2019).
Hessenberger, M. et al. Regulated membrane remodeling by Mic60 controls formation of mitochondrial crista junctions. Nat. Commun. 8, 15258 (2017).
John, G. B. et al. The mitochondrial inner membrane protein mitofilin controls cristae morphology. Mol. Biol. Cell 16, 1543–1554 (2005).
Rabl, R. et al. Formation of cristae and crista junctions in mitochondria depends on antagonism between Fcj1 and Su e/g. J. Cell Biol. 185, 1047–1063 (2009).
Tarasenko, D. et al. The MICOS component Mic60 displays a conserved membrane-bending activity that is necessary for normal cristae morphology. J. Cell Biol. 216, 889–899 (2017).
Barbot, M. et al. Mic10 oligomerizes to bend mitochondrial inner membranes at cristae junctions. Cell Metab. 21, 756–763 (2015).
Bohnert, M. et al. Central role of Mic10 in the mitochondrial contact site and cristae organizing system. Cell Metab. 21, 747–755 (2015).
Tsai, P. I., Papakyrikos, A. M., Hsieh, C. H. & Wang, X. Drosophila MIC60/mitofilin conducts dual roles in mitochondrial motility and crista structure. Mol. Biol. Cell 28, 3471–3479 (2017).
Glater, E. E., Megeath, L. J., Stowers, R. S. & Schwarz, T. L. Axonal transport of mitochondria requires milton to recruit kinesin heavy chain and is light chain independent. J. Cell Biol. 173, 545–557 (2006).
Stowers, R. S., Megeath, L. J., Gorska-Andrzejak, J., Meinertzhagen, I. A. & Schwarz, T. L. Axonal transport of mitochondria to synapses depends on milton, a novel Drosophila protein. Neuron 36, 1063–1077 (2002).
Wang, X. & Schwarz, T. L. The mechanism of Ca2+-dependent regulation of kinesin-mediated mitochondrial motility. Cell 136, 163–174 (2009).
Wang, X. et al. PINK1 and Parkin target Miro for phosphorylation and degradation to arrest mitochondrial motility. Cell 147, 893–906 (2011).
Hsieh, C. H. et al. Miro1 marks Parkinson’s disease subset and Miro1 reducer rescues neuron loss in Parkinson’s models. Cell Metab. 30, 1131–1140 (2019).
Hsieh, C. H. et al. Functional impairment in Miro degradation and mitophagy is a shared feature in familial and sporadic Parkinson’s disease. Cell Stem Cell 19, 709–724 (2016).
Shaltouki, A., Hsieh, C. H., Kim, M. J. & Wang, X. Alpha-synuclein delays mitophagy and targeting Miro rescues neuron loss in Parkinson’s models. Acta Neuropathol. 136, 607–620 (2018).
Niescier, R. F., Hong, K., Park, D. & Min, K. T. MCU interacts with Miro1 to modulate mitochondrial functions in neurons. J. Neurosci. 38, 4666–4677 (2018).
Woolums, B. M. et al. TRPV4 disrupts mitochondrial transport and causes axonal degeneration via a CaMKII-dependent elevation of intracellular Ca2+. Nat. Commun. 11, 2679 (2020).
Kornmann, B., Osman, C. & Walter, P. The conserved GTPase Gem1 regulates endoplasmic reticulum–mitochondria connections. Proc. Natl Acad. Sci. USA 108, 14151–14156 (2011).
Lee, K. S. et al. Altered ER–mitochondria contact impacts mitochondria calcium homeostasis and contributes to neurodegeneration in vivo in disease models. Proc. Natl Acad. Sci. USA 115, E8844–E8853 (2018).
Qin, J. et al. ER–mitochondria contacts promote mtDNA nucleoids active transportation via mitochondrial dynamic tubulation. Nat. Commun. 11, 4471 (2020).
Guillen-Samander, A. et al. VPS13D bridges the ER to mitochondria and peroxisomes via Miro. J. Cell Biol. https://doi.org/10.1083/jcb.202010004 (2021).
Lee, Y. J. & Chang, G. D. Quantitative display of the redox status of proteins with maleimide-polyethylene glycol tagging. Electrophoresis 40, 491–498 (2019).
Tsai, P. I. et al. PINK1 phosphorylates MIC60/Mitofilin to control structural plasticity of mitochondrial crista junctions. Mol. Cell 69, 744–756 (2018).
Markstein, M., Pitsouli, C., Villalta, C., Celniker, S. E. & Perrimon, N. Exploiting position effects and the gypsy retrovirus insulator to engineer precisely expressed transgenes. Nat. Genet. 40, 476–483 (2008).
Osterwalder, T., Yoon, K. S., White, B. H. & Keshishian, H. A conditional tissue-specific transgene expression system using inducible GAL4. Proc. Natl Acad. Sci. USA 98, 12596–12601 (2001).
Modi, S. et al. Miro clusters regulate ER–mitochondria contact sites and link cristae organization to the mitochondrial transport machinery. Nat. Commun. 10, 4399 (2019).
Tsai, P. I. et al. PINK1-mediated phosphorylation of Miro inhibits synaptic growth and protects dopaminergic neurons in Drosophila. Sci. Rep. 4, 6962 (2014).
Deng, Q. et al. Cross-talk between mitochondrial fusion and the Hippo pathway in controlling cell proliferation during Drosophila development. Genetics 203, 1777–1788 (2016).
Meng, H. et al. Loss of Parkinson’s disease-associated protein CHCHD2 affects mitochondrial crista structure and destabilizes cytochrome c. Nat. Commun. 8, 15500 (2017).
Lopez-Otin, C., Blasco, M. A., Partridge, L., Serrano, M. & Kroemer, G. The hallmarks of aging. Cell 153, 1194–1217 (2013).
Sofola, O. et al. Inhibition of GSK-3 ameliorates Abeta pathology in an adult-onset Drosophila model of Alzheimer’s disease. PLoS Genet. 6, e1001087 (2010).
Ding, W. X. et al. Nix is critical to two distinct phases of mitophagy, reactive oxygen species-mediated autophagy induction and Parkin-ubiquitin-p62-mediated mitochondrial priming. J. Biol. Chem. 285, 27879–27890 (2010).
Narendra, D., Kane, L. A., Hauser, D. N., Fearnley, I. M. & Youle, R. J. p62/SQSTM1 is required for Parkin-induced mitochondrial clustering but not mitophagy; VDAC1 is dispensable for both. Autophagy 6, 1090–1106 (2010).
Kinghorn, K. J. et al. A Drosophila model of neuronopathic Gaucher disease demonstrates lysosomal-autophagic defects and altered mTOR signalling and is functionally rescued by rapamycin. J. Neurosci. 36, 11654–11670 (2016).
Lee, V. M. & Trojanowski, J. Q. Mechanisms of Parkinson’s disease linked to pathological ɑ-synuclein: new targets for drug discovery. Neuron 52, 33–38 (2006).
Di Maio, R. et al. ɑ-Synuclein binds to TOM20 and inhibits mitochondrial protein import in Parkinson’s disease. Sci. Transl. Med. 8, 342ra378 (2016).
Ganjam, G. K. et al. Mitochondrial damage by ɑ-synuclein causes cell death in human dopaminergic neurons. Cell Death Dis. 10, 865 (2019).
Trinh, K. et al. Induction of the phase II detoxification pathway suppresses neuron loss in Drosophila models of Parkinson’s disease. J. Neurosci. 28, 465–472 (2008).
Ordonez, D. G., Lee, M. K. & Feany, M. B. ɑ-Synuclein induces mitochondrial dysfunction through spectrin and the actin cytoskeleton. Neuron 97, 108–124 (2018).
Monnier, V., Llorens, J. V. & Navarro, J. A. Impact of Drosophila models in the study and treatment of Friedreich’s ataxia. Int. J. Mol. Sci. https://doi.org/10.3390/ijms19071989 (2018).
Gottesfeld, J. M. Molecular mechanisms and therapeutics for the GAA·TTC expansion disease Friedreich ataxia. Neurotherapeutics 16, 1032–1049 (2019).
Edenharter, O., Schneuwly, S. & Navarro, J. A. Mitofusin-dependent ER stress triggers glial dysfunction and nervous system degeneration in a Drosophila model of Friedreich’s ataxia. Front. Mol. Neurosci. 11, 38 (2018).
Chen, K. et al. Loss of Frataxin induces iron toxicity, sphingolipid synthesis, and Pdk1/Mef2 activation, leading to neurodegeneration. eLife https://doi.org/10.7554/eLife.16043 (2016).
Nguyen, H. N. et al. LRRK2 mutant iPSC-derived DA neurons demonstrate increased susceptibility to oxidative stress. Cell Stem Cell 8, 267–280 (2011).
Vagnoni, A. & Bullock, S. L. A cAMP/PKA/Kinesin-1 axis promotes the axonal transport of mitochondria in aging Drosophila neurons. Curr. Biol. 28, 1265–1272 e1264 (2018).
Yang, L. et al. Multidimensional proteomics identifies declines in protein homeostasis and mitochondria as early signals for normal aging and age-associated disease in Drosophila. Mol. Cell. Proteomics 18, 2078–2088 (2019).
Menzies, F. M., Fleming, A. & Rubinsztein, D. C. Compromised autophagy and neurodegenerative diseases. Nat. Rev. Neurosci. 16, 345–357 (2015).
Lou, G. et al. Mitophagy and neuroprotection. Trends Mol. Med. 26, 8–20 (2020).
Fang, E. F. et al. Mitophagy inhibits amyloid-β and tau pathology and reverses cognitive deficits in models of Alzheimer’s disease. Nat. Neurosci. 22, 401–412 (2019).
Schuldiner, O. et al. piggyBac-based mosaic screen identifies a postmitotic function for cohesin in regulating developmental axon pruning. Dev. Cell 14, 227–238 (2008).
Guo, X. et al. The GTPase dMiro is required for axonal transport of mitochondria to Drosophila synapses. Neuron 47, 379–393 (2005).
Tricoire, H. et al. The steroid hormone receptor EcR finely modulates Drosophila lifespan during adulthood in a sex-specific manner. Mech. Ageing Dev. 130, 547–552 (2009).
Castillo-Quan, J. I. et al. Lithium promotes longevity through GSK3/NRF2-dependent hormesis. Cell Rep. 15, 638–650 (2016).
Pfeiffer, B. D. et al. Refinement of tools for targeted gene expression in Drosophila. Genetics 186, 735–755 (2010).
Ziviani, E., Tao, R. N. & Whitworth, A. J. Drosophila Parkin requires PINK1 for mitochondrial translocation and ubiquitinates Mitofusin. Proc. Natl Acad. Sci. USA 107, 5018–5023 (2010).
Castillo-Quan, J. I. et al. A triple drug combination targeting components of the nutrient-sensing network maximizes longevity. Proc. Natl Acad. Sci. USA 116, 20817–20819 (2019).
Rival, T. et al. Decreasing glutamate buffering capacity triggers oxidative stress and neuropil degeneration in the Drosophila brain. Curr. Biol. 14, 599–605 (2004).
Kinghorn, K. J. et al. Loss of PLA2G6 leads to elevated mitochondrial lipid peroxidation and mitochondrial dysfunction. Brain 138, 1801–1816 (2015).
Byers, B. et al. SNCA triplication Parkinson’s patient’s iPSC-derived DA neurons accumulate ɑ-synuclein and are susceptible to oxidative stress. PLoS ONE 6, e26159 (2011).
Kriks, S. et al. Dopamine neurons derived from human ES cells efficiently engraft in animal models of Parkinson’s disease. Nature 480, 547–551 (2011).
Wichterle, H., Lieberam, I., Porter, J. A. & Jessell, T. M. Directed differentiation of embryonic stem cells into motor neurons. Cell 110, 385–397 (2002).
Sanders, L. H. et al. LRRK2 mutations cause mitochondrial DNA damage in iPSC-derived neural cells from Parkinson’s disease patients: reversal by gene correction. Neurobiol. Dis. 62, 381–386 (2014).
Acknowledgements
We thank W. Ge, A. Whitworth and K. Zinsmaier for antibodies; L. Pallanck, W. Ge, H. Tricoire, B. Ohlstein, V. Monnier, B. Lu and Y. Imai for flies; G. Rubin and D. Rubinsztein for plasmids; Z. Tan for Miro1 protein; A. Gonzalez, R. Vanhauwaert, K. Kinghorn, J. Castillo-Quan, Y. Zhao, N. Vavouraki, J. Navarro, E. Chouchani, H. Xiao, W. Weis and the Steinberg laboratory for advice and assistance; J. Perrino and Stanford Cell Science Imaging EM Facility for support with TEM; the Vincent Coates Foundation Mass Spectrometry Laboratory at Stanford University (RRID:SCR_017801) for generating proteomic data; and the Fly Facility at the Department of Genetics of the University of Cambridge for fly injection. This work was supported by the Department of Defense (grant no. W81XWH-16-1-0282, X.W.), the NIA (grant no. R21AG06131501, X.W.), the Stanford Gabilan and McCormick Fellowship (X.W.), the Stanford Parkinson’s Disease Seed Grant (X.W.), the Stanford MCHRI Pilot Grant (X.W.), the Glenn Foundation Postdoctoral Fellowship (L.L.), Parkinson’s UK (grant no. H-1105, L.L. and L.P), and in part by the National Center for Research Resources (ARRA, grant no. 1S10RR026780-01) and the NIH (grant nos. P30CA124435 and S10RR027425).
Author information
Authors and Affiliations
Contributions
L.L., M.J.K. and A.M.P. performed the fly work. D.M.C., L.L. and V.B. were responsible for the biochemistry experiments. C.-H.H. and V.B. performed the mammalian work. A.L. and M.A.C. performed the protein purification and thermal shift assay. P.S.M., A.S.D. and K.I.A. performed the Seahorse analysis. L.P. generated the reagents and contributed to discussion. X.W. conceived and supervised the project. All authors designed the experiments and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
X.W. is a co-founder, adviser and shareholder of AcureX Therapeutics Inc., and a shareholder of Mitokinin Inc. L.L. and C.-H.H. are shareholders of AcureX Therapeutics Inc. A patent on Miro1 Reducers was filed by Stanford University with X.W., C.-H.H. and L.L. as inventors. The remaining authors declare no competing interests.
Additional information
Peer review information Primary handling editor: Isabella Samuelson. Nature Metabolism thanks Evandro Fang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Miro Binds to dMIC60 in a Redox-Dependent Manner.
(a) Two-day-old flies expressing Myc-tagged UAS-dMIC60-WT or dMIC60-CS driven by Actin-GAL4 in dMIC60 null background13,28, were lysed and immunoblotted. Both anti-dMIC60 and anti-Myc recognize transgenic dMIC60 protein in dMIC60 null background (no endogenous dMIC60)28. Anti-dMIC60 recognizes endogenous dMIC60 in flies with Actin-GAL4 alone in wild-type background. (b) Whole-body lysates of wild-type flies (w1118) were IPed with anti-DMiro or IgG, and immunoblotted (IB) as indicated. (c) Immunostaining of dMIC60 and ATP5β in third instar larval muscles. Scale bar: 5 μm. (d) Immunostaining of T7 in adult fly brains (day 7). Scale bar: 20 μm. (c-d) Confocal images were obtained using the same settings. (e) In Vitro GST pull-down using full-length GST-Miro1 or GST, together with recombinant dMIC60 (AAs 92-739). (f) In Vitro GST pull-down using full-length GST-Miro1, together with recombinant dMIC60 (AAs 92-739), either wild-type (WT) or mutant (CS). (g) Coomassie-stained gels. Arrowhead indicates the dMIC60 band. (h-i) In Vitro GST pull-down using full-length GST-Miro1 or GST, together with recombinant dMIC60 (AAs 92-739) (h) or Parkin (i). (a-i) Similar results were seen more than three times. (j) Lysates of wild-type flies fed with H2O2, paraquat, or vehicle (water) at day 7 for 24 hrs were immunoblotted as indicated. The band intensity of each marker is normalized to that of β-actin from the same blot and graphed as relative change compared to vehicle. n=4 independent experiments. Two-sided Student T Test. p=0.0336 (DMiro, H2O2), 0.0325 (dMIC60, H2O2), 0.0176 (DMiro, paraquat), 0.0383 (dMIC60, paraquat). Boxes show 25th/75th percentiles, whiskers are the minimum and maximum, and middle line is median.
Extended Data Fig. 2 DMiro Protein Levels in MICOS Mutants.
(a) Lysates of 2nd instar larvae (dMIC19null), pupae (dMIC60null), or 14-day-old adults (RNAi) were immunoblotted as indicated. The band intensity of each marker is normalized to that of ATP5β or β-actin from the same blot and graphed as relative change compared to control. Wild-type: w1118. n=3 (Top panel: anti-DMiro for WT; and Middle panel) and 4 (the rest) independent experiments. Two-sided Mann-Whitney Test (Lower panel) and T Test with Welch’s correction (Top and Middle). n.s.: not significant. p=0.0009, 0.0005, 0.0096 for dMIC19 null, 0.0319, 0.0351, 0.0173 for dMIC60 null. The rest of the precise p values are in Source Data. (b) Mitochondrial (Mito) and cytosolic (Cyto) fractions from 20-day-old flies were immunoblotted as indicated. The DMiro band intensity is normalized to that of VDAC from the same blot. n=3. Two-sided T Test with Welch’s correction. p=0.0283, 0.0102. (c) Whole-body lysates of 20-day-old flies were immunoblotted as indicated. The band intensity is normalized to that of β-actin from the same blot and graphed as relative change compared to control (Actin-GAL4, gray bar) within the same experiment. n=5 flies per experiment, 4 independent experiments. p=0.0286. (d) The mRNA levels of DMiro were determined by RT-qPCR in 20-day-old flies, normalized to RP49 from the same experiment, and graphed as relative change compared to respective controls (white boxes) within the same experiment. n=4 independent experiments. p=0.0286. (c-d) Two-sided Mann-Whitney Test. (a, d) Boxes show 25th/75th percentiles, whiskers are the minimum and maximum, and middle line is median. (a, b, c) Data are presented as Mean±S.E.M. with dots.
Extended Data Fig. 3 DMiro Protein Levels in Mitochondrial Mutants.
(a) The mRNA levels of indicated genes were determined by RT-qPCR in 20-day-old flies, normalized to RP49 from the same experiment, and graphed as relative change compared to respective controls (white boxes) within the same experiment. n=4 independent experiments. p=0.0286. (b) Whole-body lysates of 20-day-old flies were immunoblotted as indicated. The band intensity of DMiro is normalized to that of β-actin from the same blot and graphed as relative change compared to control (Actin-GAL4, gray bar). n=4 independent experiments. (c) Lysates of 2-day-old CHCHD2null and control (w1118) were immunoblotted as indicated. The band intensity of DMiro is normalized to that of β-actin from the same blot and graphed as relative change compared to wild-type control. n=4 independent experiments. (d) Lysates of 3rd instar larvae were immunoblotted. The band intensity of each marker is normalized to that of ATP5β from the same blot and graphed as relative change compared to wild-type control (w1118). n=7 larvae per experiment, 3 (anti-dMIC19 for WT) and 4 (the rest) independent experiments. p=0.0286. (e) RT-qPCR results show no significant differences in DMiro mRNA expression normalized to RP49 between young (day 3) and old (day 40) wild-type (w1118) flies. n=4 replicates per genotype. (f) Lysates of 3rd instar larvae and 2-day-old adult flies were immunoblotted with antibodies against DMiro and the loading control β-actin. The band intensity of DMiro is normalized to that of β-actin from the same blot. n=4 independent experiments. Please note that DMiro is upregulated in 2-day-old flies compared to third instar larvae. p=0.0072, 0.0047. The rest of the precise p values are in Source Data. (a) One-sided Mann-Whitney Test (dMIC19). (a-e) Two-sided Mann-Whitney Test. (f) One-Way ANOVA Post Hoc Tukey Test. (a) Boxes show 25th/75th percentiles, whiskers are the minimum and maximum, and middle line is median. (b-f) Data are presented as Mean±S.E.M. with dots.
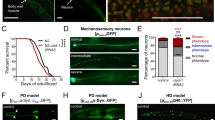
Extended Data Fig. 4 The Role of DMiro in Lifespan.
For all panels, ‘RU-‘ flies were fed with the same volume of the vehicle, ethanol. All flies were female. (a) Survival curves of wild-type flies (w1118) fed with or without 200 μM RU. n=141 flies. p=0.46 by Log-Rank Test. (b) Later life-onset neuronal downregulation of DMiro using Elav-GS-GAL4 induced in the presence of RU from day 30 (Elav-GS>UAS-DMiroRNAi, RU+, blue line) extends lifespan, compared to un-induced controls (RU-, black line). n=106 (RU+) and 127 (RU-) flies. Median lifespan was increased by 10%, p=1.3×10-7 by Log-Rank Test. (c) Adult-onset downregulation of DMiro using the intestine enterocytes driver 5966-GS-GAL4 induced in the presence of RU (5966-GS>UAS-DMiroRNAi, RU+, red line) shortens lifespan, compared to un-induced controls (RU-, black line). n=142 (RU+) and 151 (RU-) flies. p=0.0007 by Log-Rank Test. (d) Later life-onset downregulation of DMiro in intestine enterocytes using 5966-GS-GAL4 induced in the presence of RU from day 30 (5966-GS>UAS-DMiroRNAi, RU+, red line) slightly extends lifespan, compared to un-induced controls (RU-, black line). n=135 (RU-) and 145 (RU+) flies. p=0.03 by Log-Rank Test. (e) Adult-onset ubiquitous knockdown of DMiro using Da-GS-GAL4 in the presence of RU (Da-GS>UAS-DMiroRNAi, RU+, red line) shortens lifespan, compared to un-induced controls (RU-, black line). n=143 (RU+) and 144 (RU-) flies. p=2.07×10-9 by Log-Rank Test. (f) Later life-onset ubiquitous knockdown of DMiro using Da-GS-GAL4 in the presence of RU from day 30 (Da-GS>UAS-DMiroRNAi, RU+, red line) does not alter lifespan, compared to un-induced controls (RU-, black line). n=141 (RU-) and 146 (RU+) flies. p=0.74 by Log-Rank Test. (g) Adult-onset downregulation of DMiro using the muscle driver MHC-GS-GAL4 induced in the presence of RU (MHC-GS>UAS-DMiroRNAi, RU+, red line) shortens lifespan, compared to un-induced controls (RU-, black line). n=146 (RU-) and 149 (RU+) flies. p=1.68×10-16 by Log-Rank Test. (h) Adult-onset neuronal downregulation of white using Elav-GS-GAL4 induced in the presence of RU (Elav-GS>UAS-whiteRNAi, RU+, red line) slightly shortens lifespan, compared to un-induced controls (RU-, black line). n=139 flies. p=0.01 by Log-Rank Test.
Extended Data Fig. 5 dMIC60 or dMIC19 RNAi Does not Affect Mitochondrial Structure.
(a) Representative TEM images of thin sections on body wall muscles of late third instar larvae. Scale bars: 500 nm. (b) Quantification of the crista junction (CJ) number per μm of mitochondrial circumference as described in28. n=30 mitochondria from 3 male larvae. (c) Quantification of the mitochondrial size as described in28. n=19 mitochondria from 3 male larvae. (d) Quantification of the aspect ratio as described in28. n=20 mitochondria from 3 male larvae. One-Way Anova Post Hoc Tukey Test.
Extended Data Fig. 6 The Role of MICOS in Lifespan.
(a) Later life-onset downregulation of dMIC19 using the intestine enterocytes driver 5966-GS-GAL4 induced in the presence of RU from day 30 (5966-GS>UAS-dMIC19 RNAi, RU+, red line) does not alter lifespan, compared to un-induced controls (RU-, black line). n=143 (RU+) and 146 (RU-) flies. p=0.84 by Log-Rank Test. (b) Adult-onset downregulation of dMIC19 in intestine enterocytes induced in the presence of RU (5966-GS>UAS-dMIC19 RNAi, RU+, red line) does not alter lifespan, compared to un-induced controls (RU-, black line). n=144 (RU+) and 150 (RU-) flies. p=0.59 by Log-Rank Test. (c) Later life-onset downregulation of dMIC60 in intestine enterocytes induced in the presence of RU from day 30 (5966-GS>UAS-dMIC60 RNAi, RU+, red line) does not alter lifespan, compared to un-induced controls (RU-, black line). n=142 (RU+) and 143 (RU-) flies. p=0.99 by Log-Rank Test. (d) Adult-onset downregulation of dMIC60 in intestine enterocytes induced in the presence of RU (5966-GS>UAS-dMIC60 RNAi, RU+, red line) does not alter lifespan, compared to un-induced controls (RU-, black line). n=140 (RU+) and 147 (RU-) flies. p=0.94 by Log-Rank Test. (e) The survival ability of female flies with a second DMiro RNAi line driven by Elav-GS-GAL4 induced by RU since adulthood (Elav-GS>UAS-DMiroRNAi 2, blue line, RU+) or of un-induced control flies (black line, RU-), in response to 20 mM paraquat (starting from day 7). n=14 (RU+) and 56 (RU-) flies. p=0.016 by Log-Rank Test. (f) Fly heads were lysed and blotted as indicated. The band intensity is normalized to that of α-tubulin from the same blot. n=4. One-Way Anova Post Hoc Tukey Test (p=0.6915 for ubiquitin, p=0.5884 for p62). Boxes show 25th/75th percentiles, whiskers are the minimum and maximum, and middle line is median.
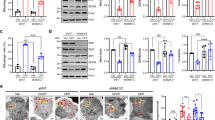
Extended Data Fig. 7 α-Syn Neurotoxicity in Flies.
(a) Validation of an α-syn fly model where wild-type human α-syn transgene downstream of a modified UAS that significantly increases gene expression59 is expressed using the inducible pan-neuronal driver Elav-GS-GAL4 (‘Elav-GS>UAS-SNCA-WT, RU+’) through adulthood. Un-induced controls are ‘Elav-GS>UAS-SNCA-WT, RU-’. Head lysates of induced and uninduced flies were immunoblotted as indicated. The band intensity of a-syn is normalized to that of β-actin. n=3 independent experiments. Two-sided T Test with Welch’s correction. p=0.0238. (b) Climbing ability shown as Performance Index of flies with adult-onset induction and of un-induced controls at different ages. n=60 (RU-) and 61 (RU+) flies per genotype, 3 independent experiments. (c) The DA neuron number in the PPM1/2, PPL1, or PPL2 clusters per brain from flies as indicated at day 40 is counted and compared. n=6 fly brains per genotype. Two-sided Mann-Whitney Test. p=0.0022, 0.0065. The rest of the precise p values are in Source Data. (d) Head lysates of 40-day-old flies were immunoblotted with antibodies against DMiro and the loading control β-actin. The band intensity of DMiro is normalized to that of β-actin from the same blot. n=3 independent experiments. Two-sided Student T Test. p=0.0138. (e) Climbing time for 30-day-old flies. n=27, 26, 57, 26 flies (from left to right). One-Way Anova Post Hoc Tukey Test. p<0.0001. (f) Flying ability of 30-day-old flies. The percentage of flies scored as a ‘1’ (able to fly) is shown in solid black bars, and the percentage of flies scored as a ‘0’ (unable to fly) is shown in white bars. n=100. Chi-Square Test. p<0.0001. (e-f) Comparisons with ‘TH-GAL4’. (g-h) HEK293T cells transfected as indicated were lysed and immunoblotted. One-Way Anova Post Hoc Tukey Test. (g) The band intensity of Miro1 is normalized to that of GAPDH from the same blot. Data from each genotype are compared to those of ‘Control RNAi, EGFP’ (the left box) and asterisk is given directly on top of the box if it is significant. Data between ‘Control RNAi, EGFP-SNCA’ and ‘MIC60 RNAi, EGFP-SNCA’ (the right 2 boxes) are compared and asterisk is given on top of a line. n=5 transfections. p=0.0004, <0.0001. The rest of the precise p values are in Source Data. (h) The band intensity of Myc-Miro1 is normalized to that of β-actin from the same blot. Data from each genotype are compared to those of ‘Myc-Miro1, dMIC60-WT, EGFP’ (the left box) and asterisk is given directly on top of the box if it is significant. Data between ‘Myc-Miro1, dMIC60-WT, EGFP-SNCA’ and ‘Myc-Miro1, dMIC60-CS, EGFP-SNCA’ (the right 2 boxes) are compared and asterisk is given on top of a line. n=5 transfections. p=0.013, 0.0006. The rest of the precise p values are in Source Data. (a, d) Data are presented as Mean±S.E.M. with dots. (c, e, g, h) Boxes show 25th/75th percentiles, whiskers are the minimum and maximum, and middle line is median.
Extended Data Fig. 8 Miro Reducers Benefit PD Models.
(a) Upper: Typical unfolding curves of human Miro1 protein in the absence (0 μM) and the presence of MR3 are shown. Lower: Quantification of melting temperatures of Miro1 protein in the absence (0 μM) and the presence of MR3 (100 μM). n=4 independent experiments. Two-sided Student T Test. p=0.0000424. (b) α-Syn-A53T-expressing flies (Elav-GAL4>UAS-SNCAA53T) were fed with 10 μM MR5 for 15 days and then their heads were lysed for immunoblotting. The band intensity of DMiro is normalized to that of the mitochondrial loading control VDAC from the same blot. n=4 independent experiments. Two-sided Mann-Whitney Test. p=0.0286. (c) Both Antimycin A and MR5 were dissolved in ethanol. Neurons were pretreated with MR5 24 hrs before the application of Antimycin A for another 6 hrs. The same volume of ethanol was applied at the same time in negative controls. Confocal images overlay triple immunostainings of TH (DA neurons), TUNEL (indicator of cell death), and Dapi (nuclei). The percentage of TUNEL-positive cells out of total cells (Dapi-positive) is calculated in each condition under 20×. n=19 (PD, MR5) or 20 fields (the rest) from 4 independent experiments. p<0.0001. The rest of the precise p values are in Source Data. (d) From images such as in (c), the percentage of TH-positive neurons out of total cells (Dapi-positive) without Antimycin A treatment ranges from 17.08%-19.25%, and is not significantly different among all conditions (p=0.9532), consistent with previous studies from ours and others20,67. n=19 (PD, MR5) or 20 fields (the rest) from 4 independent experiments. (c-d) Comparisons with ‘Wild-type, no treatment’ except otherwise indicated. One-Way Anova Post Hoc Tukey Test. Scale bar: (c) 100 µm. (a) Data are presented as Mean±S.D. with dots. (b-d) Boxes show 25th/75th percentiles, whiskers are the minimum and maximum, and middle line is median.
Supplementary information
Supplementary Table 1
Mass spectrometry data.
Supplementary Video 1
Locomotor abilities of ‘dMIC60-WT’ and ‘dMIC60-CS’ flies. Left, actin-GAL4>dMIC60-WT; dMIC60 null. Right, actin-GAL4>dMIC60-CS; dMIC60 null.
Supplementary Video 2
Flying ability of flies with TH-GAL4 alone. The flies are able to fly away immediately after being released.
Supplementary Video 3
Flying ability of a fly with expression of SNCA-A53T driven by TH-GAL4. The fly is not able to fly away even after 15 s of being released.
Supplementary Video 4
Flying ability of flies with expression of SNCA-A53T and dMIC60 RNAi driven by TH-GAL4. The flies are able to fly away either immediately or after several seconds of being released.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 1
Unprocessed western blots.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 2
Unprocessed western blots.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 6
Unprocessed western blots.
Source Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 1
Unprocessed western blots.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 2
Unprocessed western blots.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 3
Unprocessed western blots.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 6
Unprocessed western blots.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 7
Unprocessed western blots.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 8
Unprocessed western blots.
Rights and permissions
About this article
Cite this article
Li, L., Conradson, D.M., Bharat, V. et al. A mitochondrial membrane-bridging machinery mediates signal transduction of intramitochondrial oxidation. Nat Metab 3, 1242–1258 (2021). https://doi.org/10.1038/s42255-021-00443-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s42255-021-00443-2
This article is cited by
-
Mitochondrial heterogeneity in diseases
Signal Transduction and Targeted Therapy (2023)
-
Mitochondrial heterogeneity and homeostasis through the lens of a neuron
Nature Metabolism (2022)
-
MIC60 relays oxidation to block mitophagy
Nature Metabolism (2021)