Antibiotic Resistance: Finding the right sequence of drugs
In order to survive, many living organisms need to be able to adapt to their ever-changing environment. These past experiences shape the behavior of creatures big and small – from mutations in single genes to neurological changes that underlie memory formation in primates.
There is substantial evidence that environmental history affects how bacteria respond to antibiotics (Barbosa et al., 2019; Card et al., 2019; Nichol et al., 2019; Santos-Lopez et al., 2019; Yen and Papin, 2017). This has led researchers to suggest that switching between drugs over time could help slow down antibiotic resistance, as this will force the bacteria into a scenario where their existing solutions (resistance to the current drug) cannot protect them from tomorrow’s problem (a new drug). However, this method has led to mixed results (Abel zur Wiesch et al., 2014; Imamovic et al., 2018), and optimizing this approach is challenging, in part because it is unclear which features of the antibiotic sequence are the most important and guarantee the best results.
Now, in eLife, Hinrich Schulenburg (University of Kiel and Max Planck Institute for Evolutionary Biology) and colleagues – including Aditi Batra and Roderich Roemhild as joint first authors – report the results of experiments on the multi-drug resistant bacteria Pseudomonas aeruginosa (Batra et al., 2021). The team (who are based in Austria and Germany) exposed the bacteria to various sequences of three antibiotics that belong to commonly used classes of drugs: one class targets the ribosome, one targets DNA gyrase, and one targets the cell wall. In some cases, three drugs from different classes were used (that is, a heterogeneous sequence), and in some cases all three drugs belonged to the same class (a homogenous sequence). Batra et al. also varied the temporal properties of each sequence by switching between the drugs rapidly, slowly, or in a random order. The growth rate, phenotypic resistance levels and population genetics of the evolved populations were then analyzed to determine which sequences of drugs were the most effective at eliminating the bacteria (Figure 1A).
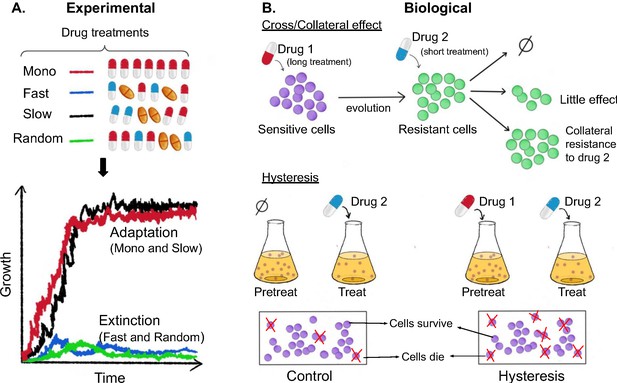
Experimental and biological features of effective drug sequences.
(A) Batra et al. applied different sequences of antibiotics to 756 populations of P. aeruginosa (top panel). The bacteria were treated with either a single drug (monotherapy; row 1), or three antibiotics which were switched rapidly (row 2), slowly (row 3) or in a random order (row 4): the three drugs were either from the same class (homogeneous) or from different classes (heterogeneous). This experiment revealed that fast (blue line) and random (green line) switching between three homogeneous beta-lactam drugs reduced bacteria growth and resulted in higher levels of extinction (bottom graph). (B) The effects of the different sequences are also impacted by biological features. (Top panel) When sensitive bacteria (shown in purple) are treated with the first drug, some cells will evolve genetic changes that make them resistant to the antibiotic treatment (shown in green). These evolutionary changes can lead to collateral effects that make the bacteria less (top arrow), equally (middle arrow) or more (bottom arrow) resistant to the second drug. (Bottom panel) Treatment with the first drug may also lead to negative hysteresis, when short-term physiological changes enhance the bacteria’s response to the second drug (right), leading to more cell death in the population compared to bacteria not pre-treated with the first drug (left).
Image credit: Anh Huynh.
In addition to these experimental parameters, the impact of different antibiotic sequences could also depend on how the population biologically responds to consecutive drug exposures (Figure 1B). For example, the genetic changes bacteria evolve in response to one antibiotic can lead to collateral effects that increase the population’s resistance or sensitivity to another drug. Because collateral resistance occurs more frequently between drugs of the same class, heterogeneous sequences of antibiotics are thought to be more effective at eliminating bacteria (Imamovic and Sommer, 2013; Lázár et al., 2013; Maltas and Wood, 2019; Pál et al., 2015; Lázár et al., 2014). Treatment with unrelated drugs has also been shown to favor negative hysteresis, which is when short-term physiological changes induced by one antibiotic enhance susceptibility towards another. Indeed, a recent study found that rapid switching between antibiotics from different classes promoted extinction of bacterial populations, even when the drugs were used at sub-inhibitory levels (Roemhild et al., 2018).
However, Batra et al. found that homogenous sequences of beta-lactams (a class of antibiotics that target the cell wall) were surprisingly more effective at clearing bacteria. The experiments also revealed that extinction tended to occur early in the treatment and was less effective when drugs were switched more slowly. A particular heterogeneous set of drugs also tended to not eliminate bacteria, indicating that heterogeneity, alone, does not guarantee success.
So, what are the important characteristics of a ‘good’ antibiotic sequence? To answer this question, Batra et al. used a common class of statistical models to probe for signatures of successful sequences. They found that extinction was strongly favored by two biological properties — low rates of spontaneous resistance and low levels of collateral resistance — and was also enhanced when switching between drugs was fast or random. This suggests that although thefindings of Batra et al. contradict the proposed benefits of using unrelated drugs, they still validate a portion of the underlying logic: using antibiotics with strong collateral effects and hysteresis enhances the impact of sequence therapy. It just turns out, however, that drugs with these characteristics are not always from different classes.
This study offers insight into how past antibiotic exposure shapes the response of bacterial populations to new challenges. In doing so, it provides a roadmap for future studies investigating how even the simplest organisms harbor signatures of past challenges and potential evolutionary solutions.
References
-
Use of collateral sensitivity networks to design drug cycling protocols that avoid resistance developmentScience Translational Medicine 5:204ra132.https://doi.org/10.1126/scitranslmed.3006609
-
Bacterial evolution of antibiotic hypersensitivityMolecular Systems Biology 9:700.https://doi.org/10.1038/msb.2013.57
-
Collateral sensitivity of antibiotic-resistant microbesTrends in Microbiology 23:401–407.https://doi.org/10.1016/j.tim.2015.02.009
Article and author information
Author details
Publication history
- Version of Record published: September 9, 2021 (version 1)
Copyright
© 2021, Huynh and Wood
This article is distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use and redistribution provided that the original author and source are credited.
Metrics
-
- 1,142
- views
-
- 82
- downloads
-
- 1
- citations
Views, downloads and citations are aggregated across all versions of this paper published by eLife.
Download links
Downloads (link to download the article as PDF)
Open citations (links to open the citations from this article in various online reference manager services)
Cite this article (links to download the citations from this article in formats compatible with various reference manager tools)
Further reading
-
- Developmental Biology
- Evolutionary Biology
Despite rapid evolution across eutherian mammals, the X-linked MIR-506 family miRNAs are located in a region flanked by two highly conserved protein-coding genes (SLITRK2 and FMR1) on the X chromosome. Intriguingly, these miRNAs are predominantly expressed in the testis, suggesting a potential role in spermatogenesis and male fertility. Here, we report that the X-linked MIR-506 family miRNAs were derived from the MER91C DNA transposons. Selective inactivation of individual miRNAs or clusters caused no discernible defects, but simultaneous ablation of five clusters containing 19 members of the MIR-506 family led to reduced male fertility in mice. Despite normal sperm counts, motility, and morphology, the KO sperm were less competitive than wild-type sperm when subjected to a polyandrous mating scheme. Transcriptomic and bioinformatic analyses revealed that these X-linked MIR-506 family miRNAs, in addition to targeting a set of conserved genes, have more targets that are critical for spermatogenesis and embryonic development during evolution. Our data suggest that the MIR-506 family miRNAs function to enhance sperm competitiveness and reproductive fitness of the male by finetuning gene expression during spermatogenesis.
-
- Evolutionary Biology
- Immunology and Inflammation
CD4+ T cell activation is driven by five-module receptor complexes. The T cell receptor (TCR) is the receptor module that binds composite surfaces of peptide antigens embedded within MHCII molecules (pMHCII). It associates with three signaling modules (CD3γε, CD3δε, and CD3ζζ) to form TCR-CD3 complexes. CD4 is the coreceptor module. It reciprocally associates with TCR-CD3-pMHCII assemblies on the outside of a CD4+ T cells and with the Src kinase, LCK, on the inside. Previously, we reported that the CD4 transmembrane GGXXG and cytoplasmic juxtamembrane (C/F)CV+C motifs found in eutherian (placental mammal) CD4 have constituent residues that evolved under purifying selection (Lee et al., 2022). Expressing mutants of these motifs together in T cell hybridomas increased CD4-LCK association but reduced CD3ζ, ZAP70, and PLCγ1 phosphorylation levels, as well as IL-2 production, in response to agonist pMHCII. Because these mutants preferentially localized CD4-LCK pairs to non-raft membrane fractions, one explanation for our results was that they impaired proximal signaling by sequestering LCK away from TCR-CD3. An alternative hypothesis is that the mutations directly impacted signaling because the motifs normally play an LCK-independent role in signaling. The goal of this study was to discriminate between these possibilities. Using T cell hybridomas, our results indicate that: intracellular CD4-LCK interactions are not necessary for pMHCII-specific signal initiation; the GGXXG and (C/F)CV+C motifs are key determinants of CD4-mediated pMHCII-specific signal amplification; the GGXXG and (C/F)CV+C motifs exert their functions independently of direct CD4-LCK association. These data provide a mechanistic explanation for why residues within these motifs are under purifying selection in jawed vertebrates. The results are also important to consider for biomimetic engineering of synthetic receptors.






