Abstract
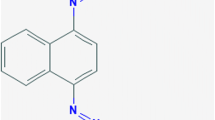
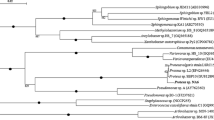
Stenotrophomonas’ metabolic versatility plays important roles in the remediation of contaminated environment and plant growth promotion. We investigated two Stenotrophomonas strains isolated from textile polluted sewage for their ability to decolorize and degrade azo dyes. Two Stenotrophomonas strains (TepeL and TepeS) were isolated from textile effluents (Tepetitla, Mexico) using the selective agar Stenotrophomonas vancomycin, imipenem, amphotericin B agar (SVIA). Isolates’ identity was determined by the sequencing of their partial 16S rRNA fragments. Their abilities to decolorize dyes were tested in a Luria broth supplemented with varying concentrations (50 mg/L–1 g/L) of textile dyes (acidic red, methyl orange, reactive green, acidic yellow, and reactive black). Fourier-transform infrared (FTIR) spectroscopy and ultra-performance liquid chromatography–mass spectrometry (UPLC-MS) metabolite analyses were used to determine the effect of the isolates’ growth on the dyes (acidic red, methyl orange). We also identified the enzymes that may be involved in the degradation process. Phylogenetic analysis based on the 16S rDNA sequences showed that the isolates belong to the genus Stenotrophomonas. Stenotrophomonas sp. TepeL and TepeS respectively decolorize all the azo dyes at the tested concentration except at 1 g/L and degraded the azo dyes. The degradation resulted in the formation of N, N-dimethyl p-phenylenediamine, and sodium 4-amino-1-naphthalenesulfonate from methyl orange and acid red. TepeL and TepeS rapidly decolorized and degraded the azo dyes tested. This result showed that the two isolates have a good potential for the decontamination of textile effluents.





Similar content being viewed by others
References
Montano S (2016) The Mexican Textile & Apparel Market representation by the US commercial Service, United States of America Department of Commerce, Dec 2016. https://otexa.trade.gov›PDFs›Textileweb16. Accessed 18th Aug 2020
World textile and apparel trade and production trends (2016) The USA, Argentina, Brazil. Colombia and Mexico Text Outlook Int 2016(179):13–50
Perkins WS (1991) A review of textile dyeing processes. Text Chem Color 23(August):23–27
Ogugbue CJ, Sawidis T (2011) Bioremediation and detoxification of synthetic wastewater containing triarylmethane dyes by aeromonas hydrophila isolated from industrial effluent. Biotechnol Res Int 2011:1–11. https://doi.org/10.4061/2011/967925
Babu BR, Parande AKRS, PKT (2016) An overview of wastes produced during cotton textile processingand effluent treatment. J Sci Cott 11(1):110–122
Khan S, Malik A (2014) Environmental and health effects of textile industry wastewater. In: Environmental deterioration and human health: natural and anthropogenic determinants. Vol 9789400778. ;:55–71. https://doi.org/10.1007/978-94-007-7890-0_4
Flores M, Manjarrez N, Solís A, Solís M, Pérez HI (2012) Microbial decolouration of azo dyes: a review. Process Biochem 47(12):1723–1748. https://doi.org/10.1016/j.procbio.2012.08.014
Chen K-C, Wu J-Y, Liou D-J, Hwang S-CJ (2003) Decolorization of the textile dyes by newly isolated bacterial strains. J Biotechnol 101(1):57–68. https://doi.org/10.1016/S0168-1656(02)00303-6
Fu Y, Viraraghavan T (2001) Fungal decolorization of dye wastewaters: A review. Bioresour Technol 79(3):251–262. https://doi.org/10.1016/S0960-8524(01)00028-1
Saratale RG, Saratale GD, Chang JS, Govindwar SP (2011) Bacterial decolorization and degradation of azo dyes: a review. J Taiwan Inst Chem Eng. Published online 2011. https://doi.org/10.1016/j.jtice.2010.06.006
Forgacs E, Cserháti T, Oros G (2004) Removal of synthetic dyes from wastewaters: a review. Environ Int 30(7):953–971. https://doi.org/10.1016/j.envint.2004.02.001
Ghoreishi SM, Haghighi R (2003) Chemical catalytic reaction and biological oxidation for treatment of non-biodegradable textile effluent. Chem Eng J 95(1–3):163–169. https://doi.org/10.1016/S1385-8947(03)00100-1
Rai HS, Bhattacharyya MS, Singh J, Bansal TK, Vats P, Banerjee UC (2005) Removal of dyes from the effluent of textile and dyestuff manufacturing industry: a review of emerging techniques with reference to biological treatment. Crit Rev Environ Sci Technol 35(3):219–238. https://doi.org/10.1080/10643380590917932
Banat IM, Nigam P, Singh D, Marchant R (1996) Microbial decolorization of textile-dye-containing effluents: a review. Bioresour Technol 58(3):217–227. https://doi.org/10.1016/S0960-8524(96)00113-7
Galai S, Limam F, Marzouki MN (2009) A new stenotrophomonas maltophilia strain producing laccase. use in decolorization of synthetics dyes. Appl Biochem Biotechnol. Published online 2009. https://doi.org/10.1007/s12010-008-8369-y
Kodam KM, Kolekar YM (2015) Bacterial degradation of textile dyes. Environ Sci Eng (Subseries Environ Sci. 2015;(9783319109411):243–266. https://doi.org/10.1007/978-3-319-10942-8_11
Khehra MS, Saini HS, Sharma DK, Chadha BS, Chimni SS (2006) Biodegradation of azo dye C.I. Acid Red 88 by an anoxic - aerobic sequential bioreactor. Dye Pigment. Published online 2006. https://doi.org/10.1016/j.dyepig.2004.12.021
Ryan RP, Monchy S, Cardinale M et al (2009) The versatility and adaptation of bacteria from the genus Stenotrophomonas. Nat Rev Microbiol 7(7):514–525. https://doi.org/10.1038/nrmicro2163
Elufisan TO, Luna ICR, Oyedara OO et al (2020) Antimicrobial susceptibility pattern of stenotrophomonas species isolated from Mexico. Afr Health Sci 20(1):168–181. https://doi.org/10.4314/ahs.v20i1.22
Elufisan TO, Rodríguez-Luna IC, Oyedara OO et al (2020) The Polycyclic Aromatic Hydrocarbon (PAH) degradation activities and genome analysis of a novel strain Stenotrophomonas sp. Pemsol isolated from Mexico PeerJ 8:e8102. https://doi.org/10.7717/peerj.8102
Benthack C, Srinivasan B, Bonvin D (2001) Partial degradation of p-aminoazobenzene by a defined mixed culture of Bacillus subtilis and Stenotrophomonas maltophilia. Biotechnol Bioeng 72(1):49–54. https://doi.org/10.1002/1097-0290(20010105)72:1%3c49::AID-BIT7%3e3.0.CO;2-X
Mohana S, Shrivastava S, Divecha J, Madamwar D (2008) Response surface methodology for optimization of medium for decolorization of textile dye Direct Black 22 by a novel bacterial consortium. Bioresour Technol 99(3):562–569. https://doi.org/10.1016/j.biortech.2006.12.033
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874. https://doi.org/10.1093/molbev/msw054
OECD (2015) Biosafety and the environmental uses of micro-organisms.; 2015. https://doi.org/10.1787/9789264213562-en
Telke AA, Kalyani DC, Dawkar VV, Govindwar SP (2009) Influence of organic and inorganic compounds on oxidoreductive decolorization of sulfonated azo dye C.I. Reactive Orange 16. J Hazard Mater 172(1):298–309. https://doi.org/10.1016/j.jhazmat.2009.07.008
Telke AA, Kalyani DC, Jadhav UU, Parshetti GK, Govindwar SP (2009) Purification and characterization of an extracellular laccase from a Pseudomonas sp. LBC1 and its application for the removal of bisphenol A. J Mol Catal B Enzym 61(3–4):252–660. https://doi.org/10.1016/j.molcatb.2009.08.001
Kerr KG, Denton M, Todd N, Corps CM, Kumari P, Hawkey PM (1996) A new selective differential medium for isolation of stenotrophomonas maltophilia. Eur J Clin Microbiol Infect Dis 15(7):607–610. https://doi.org/10.1007/BF01709373
Zhang Z, Schwartz S, Wagner L, Miller W (2000) A Greedy algorithm for aligning DNA sequences. J Comput Biol 7(1–2):203–214. https://doi.org/10.1089/10665270050081478
Kumari L, Tiwary D, Mishra PK (2016) Biodegradation of C.I. Acid Red 1 by indigenous bacteria Stenotrophomonas sp. BHUSSp X2 isolated from dye contaminated soil. Environ Sci Pollut Res 23(5):4054–0062. https://doi.org/10.1007/s11356-015-4351-8
Brooke JS (2012) Stenotrophomonas maltophilia: an emerging global opportunistic pathogen. Clin Microbiol Rev 25(1):2–41. https://doi.org/10.1128/CMR.00019-11
Denton M, Kerr KG (1998) Microbiological and clinical aspects of infection associated with Stenotrophomonas maltophilia. Clin Microbiol Rev Published online. https://doi.org/10.1074/jbc.M007003200
Aladame N (1987) Bergey’s manual of systematic bacteriology. Vol 138.; 1987. https://doi.org/10.1016/0769-2609(87)90099-8
Weber M, Schünemann W, Fuß J, Kämpfer P, Lipski A (2018) Stenotrophomonas lactitubi sp. nov. and Stenotrophomonas indicatrix sp. nov., isolated from surfaces with food contact. Int J Syst Evol Microbiol. Published online 2018. https://doi.org/10.1099/ijsem.0.002732
Sánchez-Castro I, Ruiz-Fresneda MA, Bakkali M et al (2017) Stenotrophomonas bentonitica sp. nov., isolated from bentonite formations. Int J Syst Evol Microbiol 67(8):2779–2786. https://doi.org/10.1099/ijsem.0.002016
Labat M, Thierry S, Macarie H, Cayol J-L, Ouattara AS, Assih EA (2002) Stenotrophomonas acidaminiphila sp. nov., a strictly aerobic bacterium isolated from an upflow anaerobic sludge blanket (UASB) reactor. Int J Syst Evol Microbiol. Published online 2002. https://doi.org/10.1099/00207713-52-2-559
Barragán BE, Costa C, Carmen MM (2007) Biodegradation of azo dyes by bacteria inoculated on solid media. Dye Pigment Published online. https://doi.org/10.1016/j.dyepig.2006.05.014
Ewida AYI, El-Sesy ME, Abou Zeid A (2019) Complete degradation of azo dyeacid red 337 by Bacillus megaterium KY848339.1 isolated from textile wastewater. Water Sci 33(1):154–161. https://doi.org/10.1080/11104929.2019.1688996
Eslami H, Shariatifar A, Rafiee E et al (2019) Decolorization and biodegradation of reactive Red 198 Azo dye by a new Enterococcus faecalis–Klebsiella variicola bacterial consortium isolated from textile wastewater sludge. World J Microbiol Biotechnol 35(3):38. https://doi.org/10.1007/s11274-019-2608-y
Pandey A, Singh P, Iyengar L (2007) Bacterial decolorization and degradation of azo dyes. Int Biodeterior Biodegrad 59(2):73–84. https://doi.org/10.1016/j.ibiod.2006.08.006
Franciscon E, Grossman M, Paschoal JA, Reyes FG, Durrant L (2012) Decolorization and biodegradation of reactive sulfonated azo dyes by a newly isolated Brevibacterium sp. strain VN-15. Springerplus 1:1–37. https://doi.org/10.1186/2193-1801-1-37
Zahran SA, Ali-Tammam M, Hashem AM, Aziz RK, Ali AE (2019) Azoreductase activity of dye-decolorizing bacteria isolated from the human gut microbiota. Sci Rep 9(1):5508. https://doi.org/10.1038/s41598-019-41894-8
Xie S, Huang P, Kruzic JJ, Zeng X, Qian H (2016) A highly efficient degradation mechanism of methyl orange using Fe-based metallic glass powders. Sci Rep 6(1):21947. https://doi.org/10.1038/srep21947
Parshetti G, Kalme S, Saratale G, Govindwar S. Biodegradation of malachite green by Kocuria rosea MTCC 1532. Acta Chim Slov. 2006;53(4):492–498. Accessed August 15, 2019. https://www.researchgate.net/profile/Ganesh_Saratale/publication/279903607_Biodegradation_of_malachite_green_by_Kocuria_rosea_MTCC_1532/links/55c9a27408aeca747d672738.pdf
Adnan LA, Hadibarata T, Sathishkumar P, Mohd Yusoff AR (2016) Biodegradation pathway of acid red 27 by white-rot fungus Armillaria sp. F022 and phytotoxicity evaluation. Clean - Soil, Air, Water 44(3):239–246. https://doi.org/10.1002/clen.201400249
Acknowledgements
The authors want to express their sincere appreciation to Dra Patricia Claudia Larralde Corona and Dr. Jose Alberto Narvaez Zapata of the Laboratorio de Biotecnologia Industrial for giving us the opportunity to use their sonicator.
Funding
The authors want to acknowledge CONACYT for providing the financial support for the study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declared that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Responsible Editor: Adalberto Pessoa
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Vilchis-Carmona, J.A., Rodríguez-Luna, I.C., Elufisan, T.O. et al. The decolorization and degradation of azo dyes by two Stenotrophomonas strains isolated from textile effluent (Tepetitla, Mexico). Braz J Microbiol 52, 1755–1767 (2021). https://doi.org/10.1007/s42770-021-00542-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s42770-021-00542-y