Abstract
L-lactate is an important metabolite, energy source, and signaling molecule in health and disease. In mammals, its transport across biological membranes is mediated by monocarboxylate transporters (MCTs) of the solute carrier 16 (SLC16) family. Malfunction, overexpression or absence of transporters of this family are associated with diseases such as cancer and type 2 diabetes. Moreover, lactate acts as a signaling molecule and virulence factor in certain bacterial infections. Here, we report the rational, structure-guided identification of potent, nanomolar affinity inhibitors acting on an L-lactate-specific SLC16 homologue from the bacterium Syntrophobacter fumaroxidans (SfMCT). High-resolution crystal structures of SfMCT with bound inhibitors uncovered their interaction mechanism on an atomic level and the role of water molecules in inhibitor binding. The presented systematic approach is a valuable procedure for the identification of L-lactate transport inhibitors. Furthermore, identified inhibitors represent potential tool compounds to interfere with monocarboxylate transport across biological membranes mediated by MCTs.
Similar content being viewed by others
Introduction
The solute carrier 16 (SLC16) family constitutes a diverse group of membrane transporters composed of 14 genes in the human genome (i.e., SLC16A1-A14)1. The broad range of transported substrates (e.g., L-lactate, pyruvate, ketone bodies, short-chain fatty acids, thyroid hormones, aromatic amino acids, amino acid metabolites, or drugs) reflects the diversity of the SLC16 family1. Within the transporter classification (TC) system, the SLC16 family represents the monocarboxylate transporter (MCT) family (TC-ID 2.A.1.13), which is a subgroup of the major facilitator superfamily (MFS, TC-ID 2.A.1)2. MFS transporters share a canonical fold, which is characterized by 12 transmembrane helices (TMs) assembled into two interconnected six-helix bundles3. These bundles are related to each other by a pseudo-twofold symmetry axis that runs perpendicular to the membrane plane and lies in the substrate translocation pathway. An arginine residue is conserved in transmembrane helix 8 of most SLC16 family members and its positively-charged guanidinium group is involved in the binding of the negatively-charged carboxylate group of substrates and ligands4,5,6. Several SLC16 family members are associated with health disorders. For example, MCT1 and MCT4 are overexpressed in certain tumors and play an essential role in their metabolism7. MCT11 and MCT13 are associated with type 2 diabetes risk8,9. Mutations in the MCT8 gene are connected to the Allan-Herndon-Dudley syndrome10, whereas a mutated MCT12 gene is linked to a syndrome, whose symptoms include juvenile cataract and glucosuria11. Furthermore, MCT1 deficiency is a cause of ketoacidosis12 and its failed silencing causes exercise-induced hyperinsulinism13.
L-lactate is considered an important glycolytic metabolite, energy source, and signaling molecule in human health and disease14. Additionally, lactate has an important role in some bacterial infections. In Neisseria meningitidis, it acts as a signaling molecule for microcolony dispersal15. In Staphylococcus aureus-induced biofilms, it functions as a virulence factor, sustaining an anti-inflammatory environment that promotes bacterial persistence16.
We have recently determined crystal structures of the proton-dependent, L-lactate-specific SLC16 family homologue SfMCT with bound L-lactate and thiosalicylate (TSA), which is a non-transported ligand6. The structures show SfMCT in the pharmacologically-relevant outward-open conformation and they provide important mechanistic insights into the role of critical residues involved in ligand binding6. The negatively-charged carboxylate groups of L-lactate and TSA interact with the positively-charged guanidinium group of R280 (TM8), which is conserved in TM8 of most SLC16 family members17. Here, we use this structural information for the rational identification of potent, nanomolar (nM) affinity L-lactate transport inhibitors starting from a low-affinity compound. We have solved high-resolution crystal structures of SfMCT with bound potent inhibitors to understand the molecular mechanism underlying the binding of these molecules. Furthermore, we uncovered the role of water molecules in the interaction between inhibitors and key residues based on high-quality structural data. Identified inhibitors represent potential tool compounds to interfere with monocarboxylate transport across biological membranes.
Results
Rational identification of potent L-lactate transport inhibitors
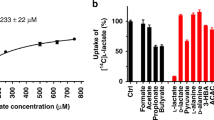
The thiosalicylate (TSA)-bound structure of the proton-dependent, L-lactate-specific SLC16 homologue SfMCT in its outward-open conformation served as a structural framework for the rational identification of potent L-lactate transport inhibitors6. The previously characterized ligand binding site provides space to accommodate molecules that are larger than TSA. Since TSA is a sulfanyl-derivative of the aromatic monocarboxylate benzoate (BA), BA was selected as starting compound of a rational search for molecules that potently inhibit L-lactate transport (Supplementary Fig. 1). Only molecules containing one carboxylate-group were considered (i.e., monocarboxylates) as R280 (TM8) is the only positively-charged residue in the ligand binding site region. Furthermore, previous experiments have shown that SfMCT does not have significant affinity for di- and tricarboxylates6. The binding site also contains hydrophobic and aromatic residues, which can be involved in hydrophobic and π-stacking interactions with bound molecules. Therefore, the focus of the screen was set on aromatic monocarboxylates, which are summarized in Supplementary Fig. 1. To evaluate the inhibitory effect of the selected molecules, an inhibition assay was set up where the transport of [14C]L-lactate through SfMCT was measured in the absence and presence of different inhibitors at decreasing concentrations. In a first step, the inhibitory effects of BA, as well as hydroxylated and methylated BA derivatives on L-lactate transport, were investigated at a concentration of 500 µM (Fig. 1A, red). Among the hydroxylated BA derivatives, a hydroxyl group at the C2-carbon atom of BA (i.e., salicylate (SA)) leads to the strongest decrease in L-lactate transport activity. BA and SA inhibit L-lactate transport with comparable Ki values of 431 µM and 362 µM, respectively (Fig. 1B, C). In contrast to hydroxylation, the C4-carbon atom of BA is the optimal position for derivatization by a methyl-group (i.e., 4-methyl-BA (4MBA)) resulting in the strongest reduction of L-lactate transport activity followed by methylation at the C3- and C2-carbon atoms (Fig. 1A, red). 4MBA inhibits L-lactate transport with a Ki of 13.4 µM (Fig. 1D), which is ~30 times more potent than BA, the starting compound of the rational search. Inhibition experiments performed at a compound concentration of 100 µM show that combining hydroxylation and methylation of BA at the determined optimal positions (i.e., C2-carbon atom for hydroxylation, C4-carbon atom for methylation) does not lead to a synergistic improvement of L-lactate transport inhibition (Fig. 1A, blue). As observed for BA derivatives, methylation at the C4-carbon atom of SA (4-methyl-SA (4MSA)) leads to the strongest reduction of L-lactate transport among the methylated SA-derivatives with a Ki of 24.4 µM (Fig. 1E). This inhibitory potency is comparable with the value obtained for 4MBA. The hydroxyl group of 4MSA cannot be shifted from the C2- to the C3-carbon atom as shown by the strongly reduced inhibition of L-lactate transport by 3-hydroxy-4-methyl-BA (3OH4MBA; Fig. 1A, blue). The fact that adding methyl-groups to the C3- and C4-carbon atoms increased the inhibitory effect of BA and SA, suggests fusing a second benzene ring to these compounds to further increase their affinity. This results in two naphthoates (i.e., 1-naphthoate (N1C) and 2-naphthoate (N2C)), and three hydroxy-naphthoate derivatives (i.e., 1-hydroxy-2-naphthoate (1OHN2C), 3-hydroxy-2-naphthoate (3OHN2C) and 2-hydroxy-1-naphthoate (2OHN1C)). The inhibitory effect of these compounds on L-lactate transport was determined at a concentration of 25 µM (Fig. 1A, orange). N2C, where the second benzene ring is fused to BA via the C3- and C4-carbon atoms, and 1OHN2C show the strongest reduction of L-lactate transport among the tested naphthoate derivatives with Ki values of 2.4 and 16.2 µM, respectively (Fig. 1F, G). 1OHN2C revealed a comparable Ki value as observed for 4MBA and 4MSA. The naphthalene rings of N2C and 1OHN2C are completely planar. In order to increase conformational flexibility, the carboxylate-containing benzene ring of N2C was replaced by an sp2 hybridized alkene (i.e., trans-cinnamate (CA)) or an sp3 hybridized alkane (i.e., 3-phenyl-propionate (3PP)) chain. CA and N2C have similar Ki values of 2.5 and 2.4 µM, respectively (Fig. 1F, H). In contrast, 3PP shows a stronger inhibitory effect compared to N2C (Fig. 1A, gray), which is highlighted by a lower Ki value of 812 nM (Fig. 1I). Accordingly, we selected aromatic monocarboxylates where the carboxylate-group and the benzene ring are connected by an aliphatic chain in the further rational screening procedure. Aromatic monocarboxylates of different length (i.e., benzoate (BA), phenylacetate (PA), 3-phenylpropionate (3PP), and 4-phenylbutyrate (4PB); Fig. 1A, black) served as molecular rulers to determine the optimal length of the aliphatic linker. Among the tested compounds, 3PP is the most potent inhibitor (Fig. 1A, black), which is ~530 times more potent than the starting compound (BA) of the rational search. In a final step, the effect on L-lactate transport of hydroxylation and methylation of the benzene ring of 3PP was systematically analyzed at an inhibitor concentration of 10 µM (Fig. 1A, magenta). Hydroxylation of the C3-carbon atom (3-(3-hydroxyphenyl)-propionate (3OH3PP)) leads to the strongest L-lactate transport reduction among the tested hydroxylated derivatives (Fig. 1A, magenta). In contrast, a methyl-group at the C2-carbon atom (3-(2-methylphenyl)-propionate (2M3PP)) reveals the strongest inhibitory effect on L-lactate transport among the tested methylated derivatives. 2M3PP inhibits L-lactate transport with a Ki value of 469 nM (Fig. 1J), which is significantly lower than the inhibition induced by 3PP. In contrast to SfMCT, the here identified key inhibitors (i.e., 1OHN2C, N2C, 3PP, and 2M3PP) do not affect the transport activity of E. coli L-lactate permeases (i.e., LldP and GlcA), whose function depends on an intact transmembrane proton-gradient (Supplementary Fig. 2)18. Therefore, the reduced SfMCT-mediated L-lactate transport, which was measured in the presence of these key inhibitors, is due to transport inhibition and does not result from a protonophoric effect of these molecules. We have also performed a transport assay using radiolabeled SA and 3PP, which are both key compounds of our rational search. SfMCT does not transport SA or 3PP, which allows the assumption that also derivatives thereof are not transported (Supplementary Fig. 3). In summary, the presented rational screening approach allowed the identification of two nM-affinity inhibitors starting from a low-affinity compound (i.e., BA). The best inhibitor 2M3PP, which has a Ki value of 469 nM, is almost 1000 times more potent than the starting compound of the rational search.
A Screening for L-lactate transport inhibitors using a transport inhibition assay (final inhibitor concentrations are indicated). Full names and molecular structures of the used compounds are given in Supplementary Fig. 1. For compounds highlighted by a star, Ki values were determined. Residual uptake in the presence of competitor is normalized with respect to control samples without competitor (control). B–J Ki determination of selected inhibitors with displayed structures. The determined Ki values and the 95% confidence intervals are indicated in the corresponding panels. Data are represented as mean ± SEM from three to five independent experiments, each in triplicate. If not visible, error bars are smaller than symbols. Individual data points are shown as open circles.
Crystal structures of SfMCT with bound inhibitors
To understand the molecular interactions underlying the binding of the identified key inhibitors (i.e., 1OHN2C, N2C, 3PP, and 2M3PP), we established a procedure to successfully co-crystallize SfMCT with these compounds. Purified and concentrated SfMCT (8 mg/ml; ~0.18 mM) was supplemented with 1 mM of key inhibitors before setting up sitting-drop crystallization trials. Crystals of sufficient size for high-quality data collection only grew in the presence of inhibitors. This allowed us to solve the crystal structures of SfMCT with bound inhibitors at resolutions of 2.39–2.67 Å (Supplementary Table 1). For all inhibitors, the transporter adopts the pharmacologically-relevant outward-open conformation, where the previously characterized binding site is accessible from the extracellular side (Fig. 2A)6. In all cases, the binding site is occupied by one inhibitor molecule (Fig. 2B–F; for OMIT maps, see Supplementary Fig. 4). Residues from the amino- and carboxy-terminal six-helix bundles interact through ionic (R280 (TM8)), hydrogen bond (Y331 (TM10), Y119 (TM4)), hydrophobic (L28 (TM1), C57 (TM2), F60 (TM2), L145 (TM5), F335 (TM10), F359 (TM11), C362 (TM11)) and π-stacking (F359 (TM11)) interactions with the inhibitors (Figs. 2 and 3). Furthermore, water molecules adopt important roles in inhibitor binding. As previously observed for L-lactate and TSA, the carboxylate-groups of all inhibitors are positioned in an ~8 Å wide confinement formed by the side chains of L145 (TM5) and F335 (TM10) (Fig. 3)6. Both oxygen atoms of the negatively-charged carboxylate-group of all inhibitors form a salt-bridge with the Nη1 and Nη2 nitrogen atoms of the positively-charged guanidinium group of the conserved and functionally important R280 (TM8). The carboxylate-groups of the bound inhibitors also interact with the hydroxyl group of Y119 (TM4) via hydrogen bonding. The orientation between the guanidinium plane of R280 (TM8) and the carboxylate-groups of bound key inhibitors was quantified by measuring their interaction geometry (Fig. 2G, H; Supplementary Fig. 5). In all cases, the angles between the guanidinium and the carboxylate planes are in a range that is commonly observed for arginine-carboxylate interactions19. In contrast, TSA interacts only with one oxygen atom of its carboxylate-group with the Nη atoms of R280 (TM8) (Fig. 2F; Supplementary Fig. 5). Furthermore, the interaction geometry is not in the range that is commonly observed for arginine-carboxylate interactions19. The naphthalene ring of 1OHN2C is almost in plane with its carboxylate-group, which can be attributed to an intramolecular hydrogen bond between the hydroxyl and carboxylate groups20. The hydroxyl group of 1OHN2C is in hydrogen bonding distance to the hydroxyl group of Y331 (TM10), which plays a role in ligand recognition6. In contrast to 1OHN2C, the naphthalene ring of N2C is rotated by ~40° with respect to the carboxylate-group. As observed for N2C, the benzene rings of 3PP and 2M3PP are also not in plane with their carboxylate-groups. The rotation of the benzene or naphthalene rings with respect to the carboxylate groups is associated with a higher conformational flexibility. This allows for a better adaptation to the binding site of SfMCT and seems to be important for improving the affinity of inhibitors.
A Overall structure of SfMCT in the outward-open conformation viewed in the plane of the membrane with indicated binding site residues (cyan) and surface representation. B–F Binding mechanism of 1OHN2C, N2C, 3PP, 2M3PP, and TSA to SfMCT. Corresponding omit maps are shown in Supplementary Fig. 4. Residues within a distance of 4 Å from the bound compounds are displayed as ball-and-stick models and highlighted in cyan. Pink spheres indicate the centers of benzene rings of aromatic residues and inhibitors, which are involved in π–π stacking interactions. Water molecules are indicated by labeled red spheres. The role of the second water molecule (2) is shown in Supplementary Fig. 7. The different rotamer conformation of L28 in the case of TSA binding is indicated by a star in (F). Distances are given in Ångström (Å). G, H The orientation between the guanidinium plane of R280 and the carboxylate-groups of bound inhibitors can be quantified by measuring the angle ψ between the Nε–Cζ and Cx–Cy bonds as well as the angle θ between the guanidinium (cyan) and the carboxylate (gray) planes. The following angles were measured: 1OHN2C ψ = 15°, θ = 15°; N2C ψ = 16°, θ = 20°; 3PP ψ = 17°, θ = 31°; 2M3PP ψ = 11°, θ = 18°; TSA ψ = 69°, θ = 16°. PDB IDs of displayed structures are 6ZGR (1OHN2C), 6ZGS (3PP), 6ZGT (N2C), 6ZGU (2M3PP) and 6G9X (TSA).
Ligands A 1OHN2C, B N2C, C 3PP, and D 2M3PP as determined by structural analysis21. Hydrophobic residues within a distance of 4 Å from the bound compounds are displayed as ball-and-stick models and highlighted in cyan. The centers of benzene rings of aromatic residues and inhibitors, which are involved in π–π stacking interactions, are indicated by pink spheres. Distances are given in Ångström (Å). E–H Surface representations of the inhibitor binding site region. Bound compounds are displayed as ball-and-stick models. A cavity between TMs 7 and 10 is indicated by a black curved line and a star. PDB IDs of displayed structures are 6ZGR (1OHN2C), 6ZGS (3PP), 6ZGT (N2C), and 6ZGU (2M3PP).
Role of Y331 and water molecules in ligand binding
An aromatic residue in TM10, which corresponds to Y331 in SfMCT, is involved in ligand recognition in several bacterial and human L-lactate transporters5,6. While the hydroxyl group of Y331 (TM10) is directly hydrogen-bonded to the hydroxyl group of 1OHN2C and the thiol group of TSA, it interacts indirectly via a water molecule with the carboxylate groups of N2C, 3PP, and 2M3PP (Fig. 2B–F). Therefore, the hydroxyl and thiol groups as well as the identified water molecule are important for the interaction with the functionally-relevant Y331 (TM10) and thus have a similar functional role in ligand binding. In addition to structural data, transport inhibition experiments corroborate the important role of Y331 (TM10) in ligand recognition. Replacing Y331 (TM10) by a phenylalanine, which removes the hydroxyl group of Y331 (TM10), results in a ~2-fold reduction of L-lactate transport inhibition by 1OHN2C, N2C, 3PP, and 2M3PP (Supplementary Fig. 6). This highlights the functional significance of the hydroxyl group of Y331 (TM10) for ligand recognition. A second water molecule was identified as part of an extended hydrogen-bonding relay network, which connects functionally-relevant residues from the amino- and carboxy-terminal six-helix bundle (i.e., L145 (TM1), N276 (TM8), R280 (TM8), and Y331 (TM10); Supplementary Fig. 7)6.
Hydrophobic interactions
In addition to salt bridges and hydrogen bonds, hydrophobic interactions contribute to inhibitor binding (Fig. 3). The benzene ring of F335 (TM10), which is part of the carboxylate group-accommodating confinement, is involved in a hydrophobic interaction with the aromatic moiety of naphthoate-derivatives, as well as with the alkane chains of 3PP and 2M3PP (Fig. 3). Furthermore, there is a hydrophobic interaction between the methyl-group of 2M3PP and the aromatic side chain of F335 (TM10), which provides an additional binding interaction with SfMCT compared to 3PP. The surface representation of the binding site region clearly highlights that the methyl-group of 2M3PP points towards a cavity between TM7 and TM10 (Fig. 3H). This cavity is exclusively located in the carboxy-terminal six-helix bundle and therefore dictates the orientation of bound 2M3PP. A hydrophobic environment formed by L28 (TM1), C57 (TM2), F60 (TM2), F359 (TM11), and C362 (TM11) accommodates the second benzene ring of the naphthalene moiety of 1OHN2C and N2C as well as the benzene ring of 3PP and 2M3PP (Fig. 3). Structural analysis revealed that the carboxylate group-containing benzene ring of N2C and the benzene rings of 3PP and 2M3PP are involved in a T-shaped π-stacking interaction with the aromatic side chain of F359 (TM11) (Fig. 3)21. With the exception of L28 (TM1), residues, which are involved in inhibitor binding, adopt similar rotamer conformations in SfMCT crystal structures with bound key inhibitors and bound TSA (Fig. 2). In contrast to the TSA-bound structure, L28 (TM1) is rotated away from the central cavity in the presence of the here identified inhibitors (Fig. 2F, star). This rotamer conformation of the isobutyl side-chain of L28 (TM1) allows the formation of hydrophobic interactions with the carboxylate group-containing benzene ring of the naphthoate-derivatives (Fig. 3A, B) or with the benzene ring of 3PP and 2M3PP (Fig. 3C, D). The distance between the benzene ring of 3PP and the adjacent TMs 2 and 11 is <4.5 Å, which precludes the extension of the inhibitor by another methylene-group. This structural information is in line with functional data. In a molecular ruler experiment using aromatic monocarboxylates, which differ in the length of the aliphatic chain connecting the carboxylate-group and the benzene ring, as L-lactate transport inhibitors (i.e., BA, PA, 3PP, and 4PB; Fig. 1A), 3PP was identified as the inhibitor with the optimal length among the tested molecules (Fig. 1A, black). Apparently, while longer molecules (i.e., 4PB) do not properly fit into the binding pocket, shorter compounds (e.g., BA, PA) cannot establish sufficient interactions with SfMCT, which reduces their affinities.
Discussion
L-lactate is no longer solely regarded as glycolytic end-product, but is now considered an important energy source and signaling molecule in human health and disease14. Also, there is evidence of its role as a virulence factor and signaling molecule during bacterial infections15,16. In human, the transport of this negatively-charged monocarboxylate across the hydrophobic core of biological membranes is mediated by members of the SLC16 family, which are homologues of the here studied SfMCT. In pathogenic bacteria, the presence of SfMCT-related lactate transporters might help promoting infections15,16. Using a rational and structure-guided screening approach with a low-affinity starting compound, we have successfully identified a series of monocarboxylates that efficiently inhibit SfMCT-mediated L-lactate transport. The identified inhibitors 3PP and 2M3PP have Ki values in the nM-range, which are up to ~1000 times lower than the one measured for the starting compound BA (Fig. 1B, I and J). The corresponding, inhibitor-bound SfMCT crystal structures highlight that the negatively-charged carboxylate groups of the inhibitors are bound to the positively-charged guanidinium group of R280 (TM8; Fig. 2) as also observed for the L-lactate- and TSA-bound structures6. The fact that the bound inhibitors share the same binding site as the transported substrate L-lactate suggests that these molecules act as competitive inhibitors. In the previously published L-lactate- and TSA-bound SfMCT crystal structures, only one oxygen atom of the carboxylate group interacted with the guanidinium group of R280 (TM8)6. This binding mode has also been observed in the substrate-bound crystal structure of the galactonate membrane transporter DgoT, which is an SLC17 family homologue22. In contrast, both carboxylate oxygen atoms of the here identified and co-crystallized inhibitors were involved in salt-bridges with this particular guanidinium group, which was also observed for carboxylate-group containing drugs bound to their targets23,24,25,26. These drugs had similar interaction geometries as determined for the here identified SfMCT inhibitors. The fact that the interaction geometries between the carboxylate groups of SfMCT inhibitors and the guanidinium group of R280 (TM8) are within a preferred range might be the reason for their lower Ki values compared to TSA19. In addition to a salt-bridge established to R280 (TM8), the carboxylate groups of the here identified inhibitors are also hydrogen-bonded to the hydroxyl group of Y119 (TM4). The combination of an interaction to an arginine and the hydroxyl group of a tyrosine was also observed for the binding of benzoate to an amino acid oxidase23 as well as for the binding of carboxylate-containing drugs to cyclooxygenases27,28,29. Our structural and functional data highlight the critical role of Y331 (TM10) in ligand binding. Whereas Y331 (TM10) is directly hydrogen-bonded to the hydroxyl group of 1OHN2C and the thiol group of TSA, it interacts indirectly via a water molecule with the carboxylate groups of N2C, 3PP, and 2M3PP (Supplementary Fig. 7). Comparison of residues that are important for inhibitor-binding in SfMCT (Figs. 2 and 3) with corresponding residues in the structure of human MCT130 indicates that there is no strong conservation except for the conserved and functionally-essential positively-charged arginine residue in TM8 (Supplementary Fig. 8). This residue is involved in ligand binding in prokaryotic and mammalian SLC16 transporters6,30,31. Alignment of the 2M3PP-bound SfMCT structure with an outward-open cryo-EM structure of human MCT130 suggests that the here presented SfMCT inhibitors (i.e., 1OHN2C, N2C, 3PP, 2M3PP) might be differently oriented in human MCT1 since the distance between the Nη atoms of the guanidinium group of R313 and the oxygen atoms of the carboxylate group of 2M3PP are 4.7 and 4.9 Å (Supplementary Fig. 8C), which is significantly larger than observed for the inhibitor-bound SfMCT structure (Fig. 2). Furthermore, there would be clashes between the inhibitors and MCT1 side chains (Supplementary Fig. 8). Nevertheless, it can be hypothesized that SfMCT inhibitors might indicate transport-modulating effects on certain human MCTs as all compounds contain a carboxylate group.
Water molecules are involved in ligand binding in several membrane transporters where they bridge the distance between transporter residues and the bound ligands32,33,34,35,36,37,38. Removing the hydroxyl group of Y331 (TM10) (i.e., Y331F mutation) reduces the transporter affinity for the identified inhibitors (Supplementary Fig. 6). This has also been previously shown for the binding of TSA6. A second water molecule is part of an extended hydrogen-bonding relay network, which connects functionally-relevant residues in the binding site region (Supplementary Fig. 7). It can be concluded that the identified water molecules contribute to the correct spatial positioning of side chains involved in ligand binding (i.e., L145 (TM5), N276 (TM8), R280 (TM8), Y331 (TM10)). The here presented SfMCT crystal structures provide a molecular explanation for the fact that 2M3PP has the best inhibitory potency among the characterized compounds. First, the sp3 hybridized alkane chain that connects the carboxylate group with the benzene ring allows a higher conformational flexibility of 3PP and 2M3PP compared to 1HN2C and N2C, which is important for efficient inhibitor binding. Second, surface analysis of all inhibitor-bound crystal structures reveals an asymmetric binding site region with a cavity in the carboxy-terminal six-helix bundle between TM7 and TM10 (Fig. 3E–H, star). The methyl group of 2M3PP points towards this cavity and dictates the orientation of bound 2M3PP, since no cavity has been identified in the amino-terminal six-helix bundle. The compound 3PP represents a promising inhibitor scaffold, whose length is mainly restricted by TMs 2, 8, and 11 (Fig. 3G). The 3PP- and 2M3PP-bound, outward-open SfMCT crystal structures suggest that the inhibitor size can be vertically increased in the direction of the substrate translocation pathway by attachments at the Cβ carbon atom (Supplementary Fig. 1).
In summary, we have identified nM-affinity transport inhibitors of the L-lactate-specific SLC16 homologue SfMCT using a rational, structure-guided screening approach, which started with a low-affinity compound. Accordingly, the inhibitory potency could be increased by almost three orders of magnitude. We have solved high-resolution crystal structures of SfMCT with bound inhibitors, which provided important insights into the binding mechanism of these molecules on an atomic level. These crystal structures clearly explain the molecular reasons, which underlie the improvement of the inhibitory potency of the identified inhibitors. This highlights the impact of high-resolution, inhibitor-bound crystal structures on the interpretation of inhibition data and the design of potent inhibitors. The here identified L-lactate transport inhibitors represent valuable compounds that might potentially also interfere with substrate transport across biological membranes mediated by SLC16 family members. In contrast to human MCTs, only a very low number of the innumerable bacterial homologs have been studied. Considering the presence of SLC16 family homologues in human pathogenic bacteria, application of monocarboxylate transport inhibitors to compromise pathogen’s survival might turn out as a valid antibacterial approach in the near future.
Methods
Cloning of SfMCT
Cloning of the bacterial SfMCT (UniProt ID code A0LNN5) gene into the pZUDF21-rbs-3C-10His plasmid was done as described previously6,39,40. The gene, which was codon-optimized for expression in E. coli (GenScript), was ligated into the pZUDF21-rbs-3C-10His plasmid using 5′-HindIII and 3′-XhoI restriction sites for overexpression, or into the pEXT20 plasmid using 5′-EcoRI and 3′-XhoI restriction sites for functional studies41. The resulting constructs (pZUDF21-rbs-SfMCT-3C-10His and pEXT20-SfMCT-3C-10His) contained recombinant SfMCT followed by a C-terminal human rhinovirus 3C (HRV3C) protease cleavage site and a decahistidine-tag (His-tag). The Y331F mutant was generated by QuikChange site-directed mutagenesis (Agilent Technologies) using the primer (5′–3′) GTT ATC GGT TGG AAC TTT GGC GCA ATG TTT ACC.
Uptake of radiolabeled L-lactate into SfMCT-expressing E. coli
Uptake experiments were done as described previously6,40. Overnight precultures of E. coli JA202 (MC4100 glcA::cat lldP::kan)42, which were transformed with a plasmid carrying SfMCT (pEXT20-SfMCT-3C-10His), were diluted 1:200 into LB-medium supplemented with 100 μg/ml ampicillin. Cultures were grown at 37 °C and 180 r.p.m. in an incubator shaker (Multitron, Infors HT). At OD600 ~0.5, isopropyl-β-D-thiogalactopyranoside was added to the culture to a final concentration of 250 µM to induce SfMCT expression. In the case of E. coli MC4100 (DSM 6574, German Collection of Microorganisms and Cell Cultures), an overnight preculture was diluted 1:200 into LB-medium supplemented with 50 μg/ml streptomycin and grown at 37 °C and 180 r.p.m. in an incubator shaker (Multitron, Infors HT). Bacteria were harvested 4 h post-induction (E. coli JA202) or 6 h post-inoculation (E. coli MC4100) (5200 × g, 10 min, room temperature). The supernatant was discarded and the bacteria were gently resuspended in uptake buffer (20 mM Bis-Tris propane-HCl (pH 6.7), 250 mM KCl). The density of the bacteria suspension was adjusted to OD600 12. Transport experiments were done in a reaction volume of 50 µl, which consisted of 20 µl cell-suspension (2.4 × 108 bacteria), 10 µl substrate master mix (67 µM sodium L-lactate spiked with [14C(U)] L-lactic acid sodium salt ([14C]L-lactate, American Radiolabeled Chemicals) to a specific activity of 0.15 Ci/mmol) and 20 µl monocarboxylate inhibitor solution. In the case of transport experiments using L-lactate, salicylate, and 3-phenylpropionate for comparison, the reaction volume of 50 µl consisted of 20 µl cell-suspension (2.4 × 108 bacteria) and 30 µl substrate master mix (67 µM sodium L-lactate spiked with [14C(U)] L-lactic acid sodium salt ([14C]L-lactate, American Radiolabeled Chemicals), or 67 µM salicylic acid spiked with [7–14C] salicylic acid ([14C]salicylic acid, American Radiolabeled Chemicals), or 67 µM 3-phenyl propionic acid spiked with [1–14C] 3-phenyl propionic acid ([14C]3-phenyl propionic acid, American Radiolabeled Chemicals) to an activity per reaction of 0.167 µCi, resulting in a final substrate concentration of 40 µM and a final activity per reaction of 0.1 µCi. All monocarboxylate inhibitors were purchased from Merck (i.e., SigmaAldrich) with the exception of 6-methyl-salicylate (6MSA) that was purchased from Fluorochem. Uptake experiments were performed in 2 ml reaction tubes (Eppendorf) at 30 °C under agitation (1000 r.p.m., Thermomixer compact, Eppendorf). After 30 min, reactions were stopped by adding 900 μl stop buffer (20 mM HEPES-NaOH (pH 7.5), 150 mM NaCl) followed by centrifugation (21,000 × g, 4 min, room temperature). Pelleted bacteria were then washed two times with 900 μl stop buffer to remove free radioligand. Finally, bacteria were lysed using 50 μl 5% (w/v) sodium dodecylsulfate and transferred into a 96-well plate (Optiplate, PerkinElmer). 150 µl scintillation cocktail (MicroScint 40, PerkinElmer) were added to each well before measuring each well for 2 min with a scintillation counter (TopCount NXT, PerkinElmer). For data analysis Prism6 (GraphPad Software) was used.
Expression of SfMCT in E. coli and membrane preparation
For crystallization experiments, SfMCT was expressed in E. coli BL21(DE3)pLysS grown at 37 °C in Luria Bertani (LB) medium supplemented with antibiotics (100 μg/ml ampicillin and 36 μg/ml chloramphenicol) at 180 r.p.m. in an incubator shaker (Multitron, Infors HT). At OD600 ~0.9, expression of SfMCT was induced by adding isopropyl-β-D-thiogalactopyranoside to a final concentration of 250 µM. After four hours, cells were harvested by centrifugation (10,000 × g, 6 min, 4 °C), resuspended in lysis buffer (45 mM Tris-HCl (pH 8), 450 mM NaCl, 4 °C) and pelleted again (10,000 × g, 25 min, 4 °C). The final cell pellet was resuspended in lysis buffer and the bacteria were lysed using an M-110P Microfluidizer (Microfluidics) operated at 1,500 bar. Low-speed centrifugation (10,000 × g, 10 min, 4 °C) was done to remove unlysed bacteria. Bacterial membranes were isolated by subjecting the supernatant of the low-speed centrifugation step to ultracentrifugation (200,000 × g, 90 min, 4 °C). The pellet was resuspended in lysis buffer, homogenized and subjected to ultracentrifugation (200,000 × g, 90 min, 4 °C). Finally, membranes were resuspended and homogenized in buffer (20 mM Tris-HCl (pH 8), 150 mM NaCl, 10% (v/v) glycerol) at 100 mg/ml and stored at −80 °C. Homogenization of the membranes was done using a glass teflon homogenizer (Sartorius).
Purification of SfMCT
Purification of SfMCT was done at 4 °C. Resuspended membranes were solubilized by gentle stirring in solubilization buffer (20 mM Tris-HCl (pH 8), 150 mM NaCl, 10% (v/v) glycerol, 4% (w/v) n-nonyl β-D-glucopyranoside (NG, Glycon Biochemicals GmbH)) for 2 h followed by ultracentrifugation (200,000 × g, 30 min, 4 °C) to remove unsolubilized material. The supernatant was diluted 1:1 with detergent-free solubilization buffer supplemented with 5 mM L-histidine. The solubilized material was then incubated with nickel-nitrilotriacetate resin (Ni-NTA; ProteinIso) (1 ml resin bed volume for solubilized membranes from one-liter expression culture) under gentle stirring for 2 h. The resin was then transferred into a column and washed with 25 column volumes of washing buffer 1 (20 mM Tris-HCl (pH 8), 150 mM NaCl, 5% (v/v) glycerol, 5 mM L-histidine, 0.4% (w/v) NG) and 25 column volumes of washing buffer 2 (20 mM Tris-HCl (pH 7), 100 mM NaCl, 0.4% (w/v) NG). SfMCT was proteolytically eluted from the column by incubating the resin with His-tagged HRV3C on a rotational shaker (~16 h)43. Eluted, undigested SfMCT and co-eluted HRV3C were removed by an additional Ni-NTA purification step.
Crystallization
Purified SfMCT was concentrated to 24 mg/ml using a 50,000 Da molecular weight cut-off concentration device (SARTORIUS Stedim Biotech, Vivaspin 2). Aggregated protein was removed by ultracentrifugation (150,000 × g, 30 min, 4 °C). Concentrated SfMCT was diluted to 8 mg/ml (~180 µM) using washing buffer 2 and supplemented with 1 mM monocarboxylate inhibitor from a 10 mM stock solution (10 mM monocarboxylate, 20 mM Tris-HCl (pH 7), 100 mM NaCl, 0.4% (w/v) NG). The diluted and supplemented SfMCT was incubated on ice for 1 h. The protein was crystallized in the sitting-drop vapor-diffusion method by mixing concentrated protein with reservoir solution (50 mM Tris-HCl (pH 7), 0.5 mM ZnBr2, 32% (v/v) PEG-1000) using a Mosquito Crystal Robot (TTP Labtech). Crystals appeared after one day of incubation at 18 °C and reached maximal size after one week. Crystals were then collected and flash frozen in liquid ethane44. Before X-ray analysis, crystals were stored in liquid nitrogen.
Data collection and structure determination
All datasets were collected on frozen crystals at the X06SA (PXI) beamline of the Swiss Light Source (SLS; Paul Scherrer Institute, Villigen, Switzerland) using an EIGER 16M detector (Dectris). The datasets were indexed and integrated with XDS45 and then merged using BLEND46 of the CCP4 program suite47 without truncation of the resolution. Scaling and averaging of symmetry-related intensities for all datasets were performed by aP_scale48 with truncation of the data at the best high-resolution along h, k or l axis determined by AIMLESS49. Due to the anisotropic nature of the diffraction data the STARANISO software (http://staraniso.globalphasing.org/) was applied. This program performs an anisotropic cut-off of merged intensity data to perform Bayesian estimation of structure amplitudes and to apply an anisotropic correction to the data. The structures were obtained by molecular replacement with Phaser50 using the SfMCT structure (PDB ID 6G9X) or the N2C structure (i.e., 2-naphthoate (N2C)-bound SfMCT structure) as search model. The final structures were obtained after multiple rounds of model building with Coot51 and refinement with phenix.refine52. The following settings were used for all refinement runs: XYZ coordinates, individual B-factors, occupancies, and TLS strategies. Full data collection, processing, and refinement statistics can be found in Supplementary Table 1. Figures involving structures were prepared using PyMol (The PyMol Molecular Graphics System; Schrödinger).
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Relevant data are available from the corresponding author on reasonable request. The atomic coordinates of inhibitor-bound SfMCT structures were deposited in the Protein Data Bank under the accession numbers: 6ZGR (1OHN2C), 6ZGS (3PP), 6ZGT (N2C), and 6ZGU (2M3PP).
References
Bosshart, P. D., Charles, R.-P., Garibsingh, R.-A. A., Schlessinger, A. & Fotiadis, D. SLC16 family: from atomic structure to human disease. Trends Biochem. Sci. 46, 28–40 (2021).
Saier, M. H. Jr. et al. The transporter classification database (TCDB): recent advances. Nucleic Acids Res. 44, D372–D379 (2016).
Bosshart, P. D. & Fotiadis, D. Secondary active transporters. Subcell. Biochem. 92, 275–299 (2019).
Price, N. T., Jackson, V. N. & Halestrap, A. P. Cloning and sequencing of four new mammalian monocarboxylate transporter (MCT) homologues confirms the existence of a transporter family with an ancient past. Biochem. J. 329, 321–328 (1998).
Rahman, B., Schneider, H., Bro, A., Deitmer, J. W. & Bro, S. Helix 8 and helix 10 are involved in substrate recognition in the rat monocarboxylate transporter MCT1. Biochemistry 38, 11577–11584 (1999).
Bosshart, P. D., Kalbermatter, D., Bonetti, S. & Fotiadis, D. Mechanistic basis of L-lactate transport in the SLC16 solute carrier family. Nat. Commun. 10, 2649 (2019).
Payen, V. L., Mina, E., Van Hée, V. F., Porporato, P. E. & Sonveaux, P. Monocarboxylate transporters in cancer. Mol. Metab. 33, 48–66 (2020).
Hara, K. et al. Genome-wide association study identifies three novel loci for type 2 diabetes. Hum. Mol. Genet. 23, 239–246 (2014).
Williams, A. L. et al. Sequence variants in SLC16A11 are a common risk factor for type 2 diabetes in Mexico. Nature 506, 97–101 (2014).
Schwartz, C. E. et al. Allan-Herndon-Dudley syndrome and the monocarboxylate transporter 8 (MCT8) gene. Am. J. Hum. Genet. 77, 41–53 (2005).
Kloeckener-Gruissem, B. et al. Mutation of solute carrier SLC16A12 associates with a syndrome combining juvenile cataract with microcornea and renal glucosuria. Am. J. Hum. Genet. 82, 772–779 (2008).
van Hasselt, P. M. et al. Monocarboxylate transporter 1 deficiency and ketone utilization. N. Engl. J. Med. 371, 1900–1907 (2014).
Otonkoski, T. et al. Physical exercise–induced hypoglycemia caused by failed silencing of monocarboxylate transporter 1 in pancreatic β cells. Am. J. Hum. Genet. 81, 467–474 (2007).
Sun, S., Li, H., Chen, J. & Qian, Q. Lactic acid: no longer an inert and end-product of glycolysis. Physiology 32, 453–463 (2017).
Sigurlásdóttir, S. et al. Host cell-derived lactate functions as an effector molecule in Neisseria meningitidis microcolony dispersal. PLoS Pathog. 13, e1006251 (2017).
Heim, C. E. et al. Lactate production by Staphylococcus aureus biofilm inhibits HDAC11 to reprogramme the host immune response during persistent infection. Nat. Microbiol. 5, 1271–1284 (2020).
Halestrap, A. P. The SLC16 gene family – Structure, role and regulation in health and disease. Mol. Asp. Med. 34, 337–349 (2013).
Núñez, M. F. et al. Transport of L-lactate, D-lactate, and glycolate by the LldP and GlcA membrane carriers of Escherichia coli. Biochem. Biophys. Res. Commun. 290, 824–829 (2002).
Armstrong, C. T., Mason, P. E., Anderson, J. L. R. & Dempsey, C. E. Arginine side chain interactions and the role of arginine as a gating charge carrier in voltage sensitive ion channels. Sci. Rep. 6, 21759 (2016).
Cochran, W. The crystal structure of salicylic acid. Acta Crystallogr. 4, 376–377 (1951).
Salentin, S., Schreiber, S., Haupt, V. J., Adasme, M. F. & Schroeder, M. PLIP: fully automated protein–ligand interaction profiler. Nucleic Acids Res. 43, W443–W447 (2015).
Leano, J. B. et al. Structures suggest a mechanism for energy coupling by a family of organic anion transporters. PLOS Biol. 17, e3000260 (2019).
Mattevi, A. et al. Crystal structure of D-amino acid oxidase: a case of active site mirror-image convergent evolution with flavocytochrome b2. Proc. Natl Acad. Sci. USA 93, 7496–7501 (1996).
Smith, B. J., Colman, P. M., Von Itzstein, M., Danylec, B. & Varghese, J. N. Analysis of inhibitor binding in influenza virus neuraminidase. Protein Sci. 10, 689–696 (2001).
Pebay-Peyroula, E. et al. Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside. Nature 426, 39–44 (2003).
Canul-Tec, J. C. et al. Structure and allosteric inhibition of excitatory amino acid transporter 1. Nature 544, 446–451 (2017).
Selinsky, B. S., Gupta, K., Sharkey, C. T. & Loll, P. J. Structural analysis of NSAID binding by prostaglandin H2 synthase: Time-dependent and time-independent inhibitors elicit identical enzyme conformations. Biochemistry 40, 5172–5180 (2001).
Gupta, K., Selinsky, B. S., Kaub, C. J., Katz, A. K. & Loll, P. J. The 2.0 Å resolution crystal structure of prostaglandin H2 synthase-1: structural insights into an unusual peroxidase. J. Mol. Biol. 335, 503–518 (2004).
Orlando, B. J., Lucido, M. J. & Malkowski, M. G. The structure of ibuprofen bound to cyclooxygenase-2. J. Struct. Biol. 189, 62–66 (2015).
Wang, N. et al. Structural basis of human monocarboxylate transporter 1 inhibition by anti-cancer drug candidates. Cell 184, 370–383.e13 (2021).
Zhang, B. et al. Cooperative transport mechanism of human monocarboxylate transporter 2. Nat. Commun. 11, 2429 (2020).
Hu, N.-J., Iwata, S., Cameron, A. D. & Drew, D. Crystal structure of a bacterial homologue of the bile acid sodium symporter ASBT. Nature 478, 408–411 (2011).
Sun, L. et al. Crystal structure of a bacterial homologue of glucose transporters GLUT1–4. Nature 490, 361–366 (2012).
Ilgü, H. et al. Insights into the molecular basis for substrate binding and specificity of the wild-type L-arginine/agmatine antiporter AdiC. Proc. Natl Acad. Sci. USA 113, 10358–10363 (2016).
Martinez Molledo, M., Quistgaard, E. M., Flayhan, A., Pieprzyk, J. & Löw, C. Multispecific substrate recognition in a proton-dependent oligopeptide transporter. Structure 26, 467–476 (2018).
Jungnickel, K. E. J., Parker, J. L. & Newstead, S. Structural basis for amino acid transport by the CAT family of SLC7 transporters. Nat. Commun. 9, 550 (2018).
Minhas, G. S. & Newstead, S. Structural basis for prodrug recognition by the SLC15 family of proton-coupled peptide transporters. Proc. Natl Acad. Sci. USA. 116, 804–809 (2019).
Bräuer, P. et al. Structural basis for pH-dependent retrieval of ER proteins from the Golgi by the KDEL receptor. Science 363, 1103–1107 (2019).
Ilgü, H. et al. Variation of the detergent-binding capacity and phospholipid content of membrane proteins when purified in different detergents. Biophys. J. 106, 1660–1670 (2014).
Bonetti, S., Hirschi, S. & Bosshart, P. D. Expression, purification and crystallization of an SLC16 monocarboxylate transporter family homologue specific for L-lactate. Protein Expr. Purif. 165, 105484 (2020).
Dykxhoorn, D. M., St. Pierre, R. & Linn, T. A set of compatible tac promoter expression vectors. Gene 177, 133–136 (1996).
Núñez, M. F., Pellicer, M. T., Badı́a, J., Aguilar, J. & Baldomà, L. The gene yghK linked to the glc operon of Escherichia coli encodes a permease for glycolate that is structurally and functionally similar to L-lactate permease. Microbiology 147, 1069–1077 (2001).
Hirschi, S. & Fotiadis, D. Purification of membrane proteins by affinity chromatography with on-column protease cleavage. Methods Mol. Biol. 2127, 139–150 (2020).
Jeckelmann, J.-M., Ilgü, H., Bosshart, P. D. & Fotiadis, D. A procedure and double-chambered device for macromolecular crystal flash-cooling in different cryogenic liquids. PLoS ONE 15, e0239706 (2020).
Kabsch, W. XDS. Acta Crystallogr. Sect. D. Biol. Crystallogr. 66, 125–132 (2010).
Foadi, J. et al. Clustering procedures for the optimal selection of data sets from multiple crystals in macromolecular crystallography. Acta Crystallogr. Sect. D. Biol. Crystallogr. 69, 1617–1632 (2013).
Winn, M. D. et al. Overview of the CCP4 suite and current developments. Acta Crystallogr. Sect. D. Biol. Crystallogr. 67, 235–242 (2011).
Vonrhein, C. et al. Data processing and analysis with the autoPROC toolbox. Acta Crystallogr. Sect. D. Biol. Crystallogr. 67, 293–302 (2011).
Evans, P. R. & Murshudov, G. N. How good are my data and what is the resolution? Acta Crystallogr. Sect. D. Biol. Crystallogr. 69, 1204–1214 (2013).
McCoy, A. J. et al. Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007).
Emsley, P., Lohkamp, B., Scott, W. G. & Cowtan, K. Features and development of Coot. Acta Crystallogr. Sect. D. Biol. Crystallogr. 66, 486–501 (2010).
Afonine, P. V. et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr. Sect. D. Biol. Crystallogr. 68, 352–367 (2012).
Acknowledgements
We thank Laura Baldomà (University of Barcelona, Spain) for providing the E. coli JA202 strain and the staff of the SLS (Paul Scherrer Institute) X06SA beamline for excellent support and advice. Financial support from the Swiss National Science Foundation SNSF (grant 310030_184980) and the NCCR TransCure is kindly acknowledged.
Author information
Authors and Affiliations
Contributions
P.D.B. and D.F. conceived and designed the experiments. P.D.B., D.K. and S.B. performed the experiments, collected and analyzed the data. P.D.B. and D.F. wrote the paper and all authors contributed to manuscript revision and approved the final version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Communications Chemistry thanks Simon Newstead and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bosshart, P.D., Kalbermatter, D., Bonetti, S. et al. The making of a potent L-lactate transport inhibitor. Commun Chem 4, 128 (2021). https://doi.org/10.1038/s42004-021-00564-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42004-021-00564-5
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.