Abstract
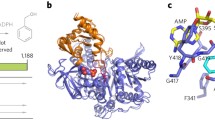
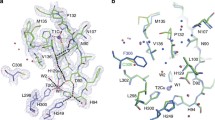
Copper nitrite reductase (CuNiR) is a copper enzyme that converts nitrite to nitric oxide and is an important part of the global nitrogen cycle in bacteria. The relatively simple CuHis3 binding site of the CuNiR active site has made it an enticing target for small molecule modeling and de novo protein design studies. We have previously reported symmetric CuNiR models within parallel three stranded coiled coil systems, with activities that span a range of three orders of magnitude. In this report, we investigate the same CuHis3 binding site within an antiparallel three helical bundle scaffold, which allows the design of asymmetric constructs. We determine that a simple CuHis3 binding site can be designed within this scaffold with enhanced activity relative to the comparable construct in parallel coiled coils. Incorporating more complex designs or repositioning this binding site can decrease this activity as much as 15 times. Comparing these constructs, we reaffirm a previous result in which a blue shift in the 1s to 4p transition energy determined by Cu(I) X-ray absorption spectroscopy is correlated with an enhanced activity within imidazole-based constructs. With this step and recent successful electron transfer site designs within this scaffold, we are one step closer to a fully functional de novo designed nitrite reductase.




Similar content being viewed by others
References
Andreini C, Bertini I, Cavallaro G, Holliday GL, Thornton JM (2008) Metal ions in biological catalysis: from enzyme databases to general principles. J Biol Inorg Chem 13(8):1205–1218
Andreini C, Bertini I, Rosato A (2009) Metalloproteomes: a Bioinformatic Approach. Acc Chem Res 42(10):1471–1479
Andreini C, Cavallaro G, Lorenzini S, Rosato A (2013) MetalPDB: a database of metal sites in biological macromolecular structures. Nucleic Acids Res 41(D1):D312–D319
Vitousek PM, Aber JD, Howarth RW, Likens GE, Matson PA, Schindler DW, Schlesinger WH, Tilman DG (1997) Human alteration of the global nitrogen cycle: sources and consequences. Ecol Appl 7(3):737–750
Galloway JN, Townsend AR, Erisman JW, Bekunda M, Cai Z, Freney JR, Martinelli LA, Seitzinger SP, Sutton MA (2008) Transformation of the nitrogen cycle: recent trends, questions, and potential solutions. Science 320(5878):889–892
Galloway JN, Leach AM, Bleeker A, Erisman JW (2013) A chronology of human understanding of the nitrogen cycle<sup>†</sup>. Philos Trans R Soc B 368(1621):20130120
Maia LB, Moura JJG (2014) How Biology Handles Nitrite. Chem Rev 114(10):5273–5357
Lehnert N, Dong HT, Harland JB, Hunt AP, White CJ (2018) Reversing nitrogen fixation. Nat Rev Chem 2(10):278–289
Iwasaki H, Noji S, Shidara S (1975) Achromobacter cycloclastes nitrite reductase the function of copper, amino acid composition, and ESR spectra1. J Biochem 78(2):355–361
Kukimoto M, Nishiyama M, Murphy MEP, Turley S, Adman ET, Horinouchi S, Beppu T (1994) X-ray Structure and Site-Directed Mutagenesis of a Nitrite Reductase from Alcaligenes Faecalis S-6: roles of two copper atoms in nitrite reduction. Biochemistry 33(17):5246–5252
Libby E, Averill BA (1992) Evidence that the Type 2 copper centers are the site of nitrite reduction by Achromobacter cycloclastes nitrite reductase. Biochem Biophys Res Commun 187(3):1529–1535
Strange RW, Dodd FE, Abraham ZHL, Grossmann JG, Brüser T, Eady RR, Smith BE, Hasnain SS (1995) The substrate-binding site in Cu nitrite reductase and its similarity to Zn carbonic anhydrase. Nat Struct Biol 2:287
Tegoni M, Yu F, Bersellini M, Penner-Hahn JE, Pecoraro VL (2012) Designing a functional type 2 copper center that has nitrite reductase activity within α-helical coiled coils. PNAS 109(52):21234–21239
Zastrow ML, Peacock AFA, Stuckey JA, Pecoraro VL (2012) Hydrolytic catalysis and structural stabilization in a designed metalloprotein. Nat Chem 4(2):118–123
Yu F, Penner-Hahn JE, Pecoraro VL, De, (2013) Novo-designed metallopeptides with type 2 copper centers: modulation of reduction potentials and nitrite reductase activities. J Am Chem Soc 135(48):18096–18107
Koebke KJ, Yu F, Salerno E, Stappen CV, Tebo AG, Penner-Hahn JE, Pecoraro VL (2018) Modifying the steric properties in the second coordination sphere of designed peptides leads to enhancement of nitrite reductase activity. Angew Chem Int Ed 57(15):3954–3957
Koebke KJ, Yu F, Van Stappen C, Pinter TBJ, Deb A, Penner-Hahn JE, Pecoraro VL (2019) Methylated histidines alter tautomeric preferences that influence the rates of cu nitrite reductase catalysis in designed peptides. J Am Chem Soc 141(19):7765–7775
Boulanger MJ, Kukimoto M, Nishiyama M, Horinouchi S, Murphy MEP (2000) Catalytic roles for two water bridged residues (Asp-98 and His-255) in the active site of copper-containing nitrite reductase. J Biol Chem 275(31):23957–23964
Shook RL, Borovik AS (2010) The role of the secondary coordination sphere in metal-mediated dioxygen activation. Inorg Chem 49(8):3646–3660
Cook SA, Borovik AS (2015) Molecular designs for controlling the local environments around metal ions. Acc Chem Res 48(8):2407–2414
Colquhoun HM, Stoddart JF, Williams DJ (1986) Second-sphere coordination—a novel rǒle for molecular receptors. Angew Chem Int Ed Engl 25(6):487–507
Koebke KJ, Alfaro VS, Pinter TBJ, Deb A, Lehnert N, Tard C, Penner-Hahn JE, Pecoraro VL (2020) Traversing the red–green–blue color spectrum in rationally designed cupredoxins. J Am Chem Soc 142(36):15282–15294
Plegaria JS, Duca M, Tard C, Friedlander TJ, Deb A, Penner-Hahn JE, Pecoraro VL, De, (2015) Novo design and characterization of copper metallopeptides inspired by native cupredoxins. Inorg Chem 54(19):9470–9482
Rittle J, Field MJ, Green MT, Tezcan FA (2019) An efficient, step-economical strategy for the design of functional metalloproteins. Nat Chem 11(5):434–441
Chino M, Leone L, Maglio O, D’Alonzo D, Pirro F, Pavone V, Nastri F, Lombardi A (2017) A de novo heterodimeric due ferri protein minimizes the release of reactive intermediates in dioxygen-dependent oxidation. Angew Chem Int Ed 56(49):15580–15583
Summa CM, Rosenblatt MM, Hong J-K, Lear JD, DeGrado WF (2002) Computational de novo design, and characterization of an A2B2 diiron protein. J Mol Biol 321(5):923–938
Marsh ENG, DeGrado WF (2002) Noncovalent self-assembly of a heterotetrameric diiron protein. PNAS 99(8):5150–5154
Cangelosi VM, Deb A, Penner-Hahn JE, Pecoraro VL (2014) A De Novo Designed Metalloenzyme for the Hydration of CO2. Angew Chem Int Ed 53(30):7900–7903
Mathieu E, Tolbert AE, Koebke KJ, Tard C, Iranzo O, Penner-Hahn JE, Policar C, Pecoraro V (2020) Rational de novo design of a cu metalloenzyme for superoxide dismutation. Chemistry 26(1):249–258
Bryson JW, Desjarlais JR, Handel TM, Degrado WF (1998) From coiled coils to small globular proteins: design of a native-like three-helix bundle. Protein Sci 7(6):1404–1414
Walsh STR, Cheng H, Bryson JW, Roder H, DeGrado WF (1999) Solution structure and dynamics of a de novo designed three-helix bundle protein. PNAS 96(10):5486–5491
Koebke KJ, Ruckthong L, Meagher JL, Mathieu E, Harland J, Deb A, Lehnert N, Policar C, Tard C, Penner-Hahn JE, Stuckey JA, Pecoraro VL (2018) Clarifying the copper coordination environment in a de novo designed red copper protein. Inorg Chem 57(19):12291–12302
Plegaria JS, Pecoraro VL (2015) Sculpting metal-binding environments in de novo designed three-helix bundles. Isr J Chem 55(1):85–95
Karayannis MI, Samios DN, Gousetis CHP (1977) A study of the molar absorptivity of ascorbic acid at different wavelengths and pH values. Anal Chim Acta 93:275–279
George, G. N.; Pickering, I. J. EXAFSPAK. http://ssrl.slac.stanford.edu/~george/exafspak/exafs.htm.
Ankudinov AL, Rehr JJ (1997) Relativistic calculations of spin-dependent x-ray-absorption spectra. Phys Rev B 56(4):R1712–R1716
Weng T-C, Waldo GS, Penner-Hahn JE (2005) A method for normalization of X-ray absorption spectra. J Synchrotron Radiat 12(4):506–510
Dimakis N, Bunker G (2004) XAFS Debye-Waller factors for Zn metalloproteins. Phys. Rev. B 70(19):195114
Pinter TBJ, Manickas EC, Tolbert AE, Koebke KJ, Deb A, Penner-Hahn JE, Pecoraro VL (2020) Making or breaking metal-dependent catalytic activity: the role of stammers in designed three-stranded coiled coils. Angew Chem Int Ed 59(46):20445–20449
Kau LS, Spira-Solomon DJ, Penner-Hahn JE, Hodgson KO, Solomon EI (1987) X-ray absorption edge determination of the oxidation state and coordination number of copper. Application to the type 3 site in Rhus vernicifera laccase and its reaction with oxygen. J Am Chem Soc 109(21):6433–6442
Chakraborty S, Kravitz JY, Thulstrup PW, Hemmingsen L, DeGrado WF, Pecoraro VL (2011) Realization of a designed three-helix bundle capable of binding heavy metals in a Tris(Cysteine) environment. Angew Chem Int Ed Engl 50(9):2049–2053
Lovejoy B, Choe S, Cascio D, McRorie D, DeGrado W, Eisenberg D (1993) Crystal structure of a synthetic triple-stranded alpha-helical bundle. Science 259(5099):1288–1293
Plegaria JS, Dzul SP, Zuiderweg ERP, Stemmler TL, Pecoraro VL (2015) Apoprotein structure and metal binding characterization of a de novo designed peptide, α3div, that sequesters toxic heavy metals. Biochemistry 54(18):2858–2873
Tebo AG, Pinter TBJ, García-Serres R, Speelman AL, Tard C, Sénéque O, Blondin G, Latour J-M, Penner-Hahn J, Lehnert N, Pecoraro VL (2018) Development of a rubredoxin-type center embedded in a de novo-designed three-helix bundle. Biochemistry 57(16):2308–2316
Polizzi NF, Wu Y, Lemmin T, Maxwell AM, Zhang S-Q, Rawson J, Beratan DN, Therien MJ, DeGrado WF, De, (2017) novo design of a hyperstable non-natural protein–ligand complex with sub-Å accuracy. Nat Chem 9(12):1157–1164
Acknowledgements
V.L.P thanks the National Institutes of Health for financial support of this research (GM141086). Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS or NIH. A.G.T. acknowledges support from the Rackham Merit Fellowship (University of Michigan), as well as training grant support from the University of Michigan Chemistry–Biology Interface (CBI) training program (NIH grant 5T32GM008597) and support from the Chateaubriand Fellowship (Embassy of France in the United States).
Funding
V.L.P thanks the National Institutes of Health for financial support of this research (GM141086). Use of the Stanford Synchrotron Radiation Lightsource, SLAC National Accelerator Laboratory, is supported by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences under Contract No. DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research, and by the National Institutes of Health, National Institute of General Medical Sciences (including P41GM103393). The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS or NIH. A.G.T. acknowledges support from the Rackham Merit Fellowship (University of Michigan), as well as training grant support from the University of Michigan Chemistry-Biology Interface (CBI) training program (NIH grant 5T32GM008597) and support from the Chateaubriand Fellowship (Embassy of France in the United States).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare no conflict of interest.
Availability of data and material
The datasets generated during and analyzed during the current study are available from the corresponding author on reasonable request.
Code availability
Not Applicable.
Supporting information
Supporting information includes a table of Cu(I) EXAFS fitting parameters and figures of the best Cu(I) EXAFS fit for each construct reported.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Koebke, K.J., Tebo, A.G., Manickas, E.C. et al. Nitrite reductase activity within an antiparallel de novo scaffold. J Biol Inorg Chem 26, 855–862 (2021). https://doi.org/10.1007/s00775-021-01889-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00775-021-01889-1