Abstract
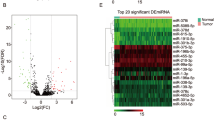
Some lncRNA-associated competing endogenous RNAs (ceRNAs) are considered as potential biomarkers for targeted therapies and prognosis in human cancer. In our present study, we aimed to construct a ceRNA network and establish a genomic-clinicopathologic nomogram to provide insights into the molecular mechanisms and predict survival for HBV-related HCC. The Cancer Genome Atlas (TCGA) database was applied to collect the data of LIHC RNA-seq dataset and miRNA-seq dataset as well as the clinicopathological information. Identification of differentially expressed RNAs (mRNAs, lncRNAs, and miRNAs) between HBV-related HCC samples and normal samples was conducted using Limma package in R. The Database for Annotation, Visualization, and Integrated Discovery (DAVID) was used for performing the functional enrichment analysis of differentially expressed mRNAs. The ceRNA network was carried out using Cytoscape. The LASSO-penalized Cox regression analysis was implemented to identify HCC-related lncRNAs, and the multivariate Cox regression analysis was conducted for the establishment of a genomic-clinicopathology nomogram. A total of 1859 DEmRNAs, 113 DElncRNAs, and 89 DEmiRNAs were screened out etween HBV-related HCC samples and normal samples. A ceRNA network including 44 DEmRNAs, 7 DElncRNAs, and 20 DEmiRNAs was constructed. 7 DElncRNAs (PVT1, LINC01138, LINC02499, AL355488.2, FGF14-AS2, MAFG-AS1 and LINC00261) were finally identified as prognostic indicators. The area under the curve reached 0.8169 for the 7-lncRNA signature. The predictive accuracy and clinical application value were remarkably high for the genomic-clinicopathologic nomogram integrating the histological grade and the 7-gene-based prognostic index. Taken together, we have established a ceRNA network with HBV-related HCC-specific DElncRNAs, DEmiRNAs, and DEmRNAs. Furthermore, the genome-wide data of lncRNA expression were analyzed using the TCGA database, and a 7-lncRNA signature was identified as a potential prognostic predictor for HBV-related HCC patients. Novel functional studies were provided by our current findings for elucidating the molecular mechanism of lncRNA in HBV-related HCC.







Similar content being viewed by others
Data availability statement
All the data supporting the conclusions of this article are included in the article and its supplementary information files.
References
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
Akamatsu N, Kokudo N. Liver transplantation for hepatocellular carcinoma from living-donor vs deceased donor. Hepatobiliary Surg Nutr. 2016;5(5):422–8.
An P, Xu J, Yu Y, Winkler CA. Host and viral genetic variation in HBV-related hepatocellular carcinoma. Front Genet. 2018;9:261.
Kalyan A, Nimeiri H, Kulik L. Systemic therapy of hepatocellular carcinoma: current and promising. Clin Liver Dis. 2015;19(2):421–32.
Finn RS, Zhu AX, Farah W, Almasri J, Zaiem F, Prokop LJ, et al. Therapies for advanced stage hepatocellular carcinoma with macrovascular invasion or metastatic disease: a systematic review and meta-analysis. Hepatology. 2018;67(1):422–35.
Chandra GS, Nandan TY. Potential of long non-coding RNAs in cancer patients: from biomarkers to therapeutic targets. Int J Cancer. 2017;140(9):1955–67.
Zhou YQ, Huan L, Wu YJ, Bao CY, Chen B, Wang L, et al. LncRNA ID2-AS1 suppresses tumor metastasis by activating the HDAC8/ID2 pathway in hepatocellular carcinoma. Cancer Lett. 2020;469:399–409.
Feng JY, Yang G, Liu YX, Gao Y, Zhao M, Bu YN, et al. LncRNA PCNAP1 modulates hepatitis B virus replication and enhances tumor growth of liver cancer. Theranostics. 2019;9(18):5227–45.
Gandhi M, Gross M, Holler JM, Coggins SA, Patil N, Leupold JH, et al. The lncRNA lincNMR regulates nucleotide metabolism via a YBX1-RRM2 axis in cancer. Nat Commun. 2020;11(1):3214.
Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. 2016;29(4):452–63.
Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP. A ceRNA hypothesis: the Rosetta Stone of a hidden RNA language? Cell. 2011;146(3):353–8.
Chen JN, Yu Y, Li H, Hu QY, Chen XL, He YT, et al. Long non-coding RNA PVT1 promotes tumor progression by regulating the miR-143/HK2 axis in gallbladder cancer. Mol Cancer. 2019;18(1):33.
Chen DL, Lu YX, Zhang JX, Wei XL, Wang F, Zeng ZL, et al. Long non-coding RNA UICLM promotes colorectal cancer liver metastasis by acting as a ceRNA for microRNA-215 to regulate ZEB2 expression. Theranostics. 2017;7(19):4836–49.
El-Serag HB. Epidemiology of viral hepatitis and hepatocellular carcinoma. Gastroenterology. 2012;142(6):1264–73.
Yang JD, Hainaut P, Gores GJ, Amadou A, Plymoth A, Roberts LR. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16(10):589–604.
Dey BK, Mueller AC, Dutta A. Long non-coding RNAs as emerging regulators of differentiation, development, and disease. Transcription. 2014;5(4):e944014.
Yin Z, Ma TT, Yan JH, Shi N, Zhang CZ, Lu X, et al. LncRNA MAGI2-AS3 inhibits hepatocellular carcinoma cell proliferation and migration by targeting the miR-374b-5p/SMG1 signaling pathway. J Cell Physiol. 2019;234(10):18825–36.
Zhang FJ, Yang C, Xing ZY, Liu P, Zhang B, Ma X, et al. LncRNA GAS5 mediated miR-1323 promotes tumor progression by targeting TP53INP1 in hepatocellular carcinoma. Onco Targets Ther. 2019;12:4013–23.
Tu WH, Yang YH, Song YL, Zhu WJ. Hepatitis B virus x protein accelerated the proliferation of hepatocellular carcinoma cell through lncRNA SNHG20/PTEN pathway. J Biochem. 2019;165(5):423–31.
Lopez-Lazaro M. The stem cell division theory of cancer. Crit Rev Oncol Hematol. 2018;123:95–113.
Shcherba M, Liang YX, Fernandes D, Perez-Soler R, Cheng HY. Cell cycle inhibitors for the treatment of NSCLC. Expert Opin Pharmacother. 2014;15(7):991–1004.
Dang SW, Zhou JS, Chen YJ, Chen P, Ji MJ, Shi BY, et al. Dynamic expression of ZNF382 and its tumor-suppressor role in hepatitis B virus-related hepatocellular carcinogenesis. Oncogene. 2019;38(24):4804–19.
Liu L, Dong Z, Liang J, Cao C, Sun J, Ding Y, et al. As an independent prognostic factor, FAT10 promotes hepatitis B virus-related hepatocellular carcinoma progression via Akt/GSK3beta pathway. Oncogene. 2014;33(7):909–20.
Xu J, Zhang J, Shan F, Wen J, Wang Y. SSTR5AS1 functions as a ceRNA to regulate CA2 by sponging miR15b5p for the development and prognosis of HBV related hepatocellular carcinoma. Mol Med Rep. 2019;20(6):5021–31.
Wu SX, Huang J, Liu ZW, Chen HG, Guo P, Cai QQ, et al. A genomic-clinicopathologic nomogram for the preoperative prediction of lymph node metastasis in bladder cancer. EBioMedicine. 2018;31:54–65.
Cui J, Wen QQ, Tan XJ, Chen Z, Liu GL. A genomic-clinicopathologic nomogram predicts survival for patients with laryngeal squamous cell carcinoma. Dis Markers. 2019;2019:5980567.
Li Z, Zhang JW, Liu XY, Li SL, Wang QF, Chen D, et al. The LINC01138 drives malignancies via activating arginine methyltransferase 5 in hepatocellular carcinoma. Nat Commun. 2018;9(1):1572.
Jiang B, Yang B, Wang Q, Zheng XY, Guo Y, Lu W. lncRNA PVT1 promotes hepatitis B virus-positive liver cancer progression by disturbing histone methylation on the c-Myc promoter. Oncol Rep. 2020;43(2):718–26.
Ouyang H, Zhang L, Xie Z, Ma SM. Long noncoding RNA MAFG-AS1 promotes proliferation, migration and invasion of hepatocellular carcinoma cells through downregulation of miR-6852. Exp Ther Med. 2019;18(4):2547–53.
Zhang HF, Li W, Han YD. LINC00261 suppresses cell proliferation, invasion and Notch signaling pathway in hepatocellular carcinoma. Cancer Biomark. 2018;21(3):575–82.
Ma XY, Mo ML, Tan H, Tan C, Zeng XZ, Zhang GQ, et al. LINC02499, a novel liver-specific long non-coding RNA with potential diagnostic and prognostic value, inhibits hepatocellular carcinoma cell proliferation, migration, and invasion. Hepatol Res. 2020;50(6):726–40.
Wu J, Liu LY, Jin HL, Li Q, Wang ST, Peng BG. LncSNHG3/miR-139-5p/BMI1 axis regulates proliferation, migration, and invasion in hepatocellular carcinoma. Onco Targets Ther. 2019;12:6623–38.
Liu JX, Lu CH, Xiao MB, Jiang F, Qu LS, Ni RZ. Long non-coding RNA SNHG20 predicts a poor prognosis for HCC and promotes cell invasion by regulating the epithelial-to-mesenchymal transition. Biomed Pharmacother. 2017;89:857–63.
Yang F, Liu YH, Dong SY, Ma RM, Bhandari A, Zhang XH, et al. A novel long non-coding RNA FGF14-AS2 is correlated with progression and prognosis in breast cancer. Biochem Biophys Res Commun. 2016;470(3):479–83.
Jin YC, Zhang M, Duan R, Yang JS, Yang Y, Wang J, et al. Long noncoding RNA FGF14-AS2 inhibits breast cancer metastasis by regulating the miR-370-3p/FGF14 axis. Cell Death Discov. 2020;6:103.
Acknowledgements
Thanks for all the help from my tutors and friends.
Funding
Support for this study was provided by the Fundamental Research Funds for the Central Universities (No.2042020kf0155), and the Medical Science Advancement Program (Clinical Medicine) of Wuhan University (No. TFLC2018003).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Human ethics
We comply with National Medical Products Administration/Good Clinical Practice (NMPA/GCP) Principle and Helsinki Declaration. And this study was approved from the Medical Ethics Committee of Zhongnan Hospital of Wuhan University (Numbers: 2020178).
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Huang, K., Lu, Z., Li, L. et al. Construction of a ceRNA network and a genomic-clinicopathologic nomogram to predict survival for HBV-related HCC. Human Cell 34, 1830–1842 (2021). https://doi.org/10.1007/s13577-021-00607-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13577-021-00607-y