Abstract
Background
Monoallelic variants in the KIF1A gene are associated with a large set of clinical phenotypes including neurodevelopmental and neurodegenerative disorders, underpinned by a broad spectrum of central and peripheral nervous system involvement.
Methods
In a multicenter study conducted in patients presenting spastic gait or complex neurodevelopmental disorders, we analyzed the clinical, genetic and neuroradiological features of 28 index cases harboring heterozygous variants in KIF1A. We conducted a literature systematic review with the aim to comparing our findings with previously reported KIF1A-related phenotypes.
Results
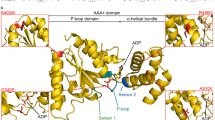
Among 28 patients, we identified nine novel monoallelic variants, and one a copy number variation encompassing KIF1A. Mutations arose de novo in most patients and were prevalently located in the motor domain. Most patients presented features of a continuum ataxia-spasticity spectrum with only five cases showing a prevalently pure spastic phenotype and six presenting congenital ataxias. Seventeen mutations occurred in the motor domain of the Kinesin-1A protein, but location of mutation did not correlate with neurological and imaging presentations. When tested in 15 patients, muscle biopsy showed oxidative metabolism alterations (6 cases), impaired respiratory chain complexes II + III activity (3/6) and low CoQ10 levels (6/9). Ubiquinol supplementation (1gr/die) was used in 6 patients with subjective benefit.
Conclusions
This study broadened our clinical, genetic, and neuroimaging knowledge of KIF1A-related disorders. Although highly heterogeneous, it seems that manifestations of ataxia-spasticity spectrum disorders seem to occur in most patients. Some patients also present secondary impairment of oxidative metabolism; in this subset, ubiquinol supplementation therapy might be appropriate.







Similar content being viewed by others
Change history
15 October 2021
A Correction to this paper has been published: https://doi.org/10.1007/s00415-021-10839-5
16 February 2023
A Correction to this paper has been published: https://doi.org/10.1007/s00415-023-11589-2
References
Blackstone C (2018) Converging cellular themes for the hereditary spastic paraplegias. Curr Opin Neurobiol 51:139–146. https://doi.org/10.1016/j.conb.2018.04.025
Okada Y, Yamazaki H, Sekine-Aizawa Y, Hirokawa Y (1995) The neuron-specific kinesin superfamily protein KIF1A is a unique monomeric motor for anterograde axonal transport of synaptic vesicle precursors. Cell 81:769–780. https://doi.org/10.1016/0092-8674(95)90538-3
Rivire JB, Ramalingam S, Lavastre V, Shekarabi M, Holbert S, Lafontaine J, Srour M, Merner N, Rochefort D, Hince P, Gaudet R, Mes-Masson AM, Baets J, Houlden H, Brais B, Nicholson GA, Van Esch H, Nafissi S, De Jonghe P, Reilly MM, Timmerman V, Dion PA, Rouleau GA (2011) KIF1A, an axonal transporter of synaptic vesicles, is mutated in hereditary sensory and autonomic neuropathy type 2. Am J Hum Genet 89:219–230. https://doi.org/10.1016/j.ajhg.2011.06.013
D’Amore A, Tessa A, Casali C, Dotti MT, Filla A, Silvestri G, Antenora A, Astrea G, Barghigiani M, Battini R, Battisti C, Bruno I, Cereda C, Dato C, Di Iorio G, Donadio V, Felicori M, Fini N, Fiorillo C, Gallone S, Gemignani F, Gigli GL, Graziano C, Guerrini R, Gurrieri F, Kariminejad A, Lieto M, Marques Lourenço C, Malandrini A, Mandich P, Marcotulli C, Mari F, Massacesi L, Melone MAB, Mignarri A, Milone R, Musumeci O, Pegoraro E, Perna A, Petrucci A, Pini A, Pochiero F, Pons MR, Ricca I, Rossi S, Seri M, Stanzial F, Tinelli F, Toscano A, Valente M, Federico A, Rubegni A, Santorelli FM (2018) Next generation molecular diagnosis of hereditary spastic paraplegias: an Italian cross-sectional study. Front Neurol 9:1–13. https://doi.org/10.3389/fneur.2018.00981
Nesti C, Meschini MC, Meunier B, Sacchini M, Doccini S, Romano A, Petrillo S, Pezzini I, Seddiki N, Rubegni A, Piemonte F, Alice Donati M, Brasseur G, Santorelli FM (2014) Additive effect of nuclear and mitochondrial mutations in a patient with mitochondrial encephalomyopathy. Hum Mol Genet 24:3248–3256. https://doi.org/10.1093/hmg/ddv078
Mero S, Salviati L, Leuzzi V, Rubegni A, Calderan C, Nardecchia F, Galatolo D, Desbats MA, Naef V, Gemignani F, Novelli M, Tessa A, Battini R, Santorelli FM, Marchese M (2021) New pathogenic variants in COQ4 cause ataxia and neurodevelopmental disorder without detectable CoQ10 deficiency in muscle or skin fibroblasts. J Neurol. https://doi.org/10.1007/s00415-021-10509-6
Nitta R, Kikkawa M, Okada Y, Hirokawa N (2004) KIF1A alternately uses two loops to bind microtubules. Science 305:678–683. https://doi.org/10.1126/science.1096621
Kikkawa M, Hirokawa N (2006) High-resolution cryo-EM maps show the nucleotide binding pocket of KIF1A in open and closed conformations. EMBO J 25:4187–4194. https://doi.org/10.1038/sj.emboj.7601299
Guerois R, Nielsen JE, Serrano L (2002) Predicting changes in the stability of proteins and protein complexes: a study of more than 1000 mutations. J Mol Biol 320:369–387. https://doi.org/10.1016/S0022-2836(02)00442-4
Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. https://doi.org/10.1002/jcc.20084
Liberati A, Altman DG, Tetzlaff J, Mulrow C, Gøtzsche PC, Ioannidis JPA, Clarke M, Devereaux PJ, Kleijnen J, Moher D (2009) The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. PLoS Med 6:e1000100. https://doi.org/10.1371/journal.pmed.1000100
Hamdan FF, Gauthier J, Araki Y, Lin DT, Yoshizawa Y, Higashi K, Park AR, Spiegelman D, Dobrzeniecka S, Piton A, Tomitori H, Daoud H, Massicotte C, Henrion E, Diallo O, Shekarabi M, Marineau C, Shevell M, Maranda B, Mitchell G, Nadeau A, D’Anjou G, Vanasse M, Srour M, Lafrenière RG, Drapeau P, Lacaille JC, Kim E, Lee JR, Igarashi K, Huganir RL, Rouleau GA, Michaud JL (2011) Excess of de novo deleterious mutations in genes associated with glutamatergic systems in nonsyndromic intellectual disability. Am J Hum Genet 88:306–316. https://doi.org/10.1016/j.ajhg.2011.02.001
Samanta D, Gokden M (2019) PEHO syndrome: KIF1A mutation and decreased activity of mitochondrial respiratory chain complex. J Clin Neurosci 61:298–301. https://doi.org/10.1016/j.jocn.2018.10.091
Okamoto N, Miya F, Tsunoda T, Yanagihara K, Kato M, Saitoh S, Yamasaki M, Kanemura Y, Kosaki K (2014) KIF1A mutation in a patient with progressive neurodegeneration. J Hum Genet 59:639–641. https://doi.org/10.1038/jhg.2014.80
Esmaeeli Nieh S, Madou MRZ, Sirajuddin M, Fregeau B, McKnight D, Lexa K, Strober J, Spaeth C, Hallinan BE, Smaoui N, Pappas JG, Burrow TA, McDonald MT, Latibashvili M, Leshinsky-Silver E, Lev D, Blumkin L, Vale RD, Barkovich AJ, Sherr EH (2015) De novo mutations in KIF1A cause progressive encephalopathy and brain atrophy. Ann Clin Transl Neurol 2:623–635. https://doi.org/10.1002/acn3.198
Lee JR, Srour M, Kim D, Hamdan FF, Lim SH, Brunel-Guitton C, Décarie JC, Rossignol E, Mitchell GA, Schreiber A, Moran R, Van Haren K, Richardson R, Nicolai J, Oberndorff KMEJ, Wagner JD, Boycott KM, Rahikkala E, Junna N, Tyynismaa H, Cuppen I, Verbeek NE, Stumpel CTRM, Willemsen MA, de Munnik SA, Rouleau GA, Kim E, Kamsteeg EJ, Kleefstra T, Michaud JL (2015) De novo mutations in the motor domain of KIF1A cause cognitive impairment, spastic paraparesis, axonal neuropathy, and cerebellar atrophy. Hum Mutat 36:69–78. https://doi.org/10.1002/humu.22709
Ohba C, Haginoya K, Osaka H, Kubota K, Ishiyama A, Hiraide T, Komaki H, Sasaki M, Miyatake S, Nakashima M, Tsurusaki Y, Miyake N, Tanaka F, Saitsu H, Matsumoto N (2015) De novo KIF1A mutations cause intellectual deficit, cerebellar atrophy, lower limb spasticity and visual disturbance. J Hum Genet 60:739–742. https://doi.org/10.1038/jhg.2015.108
Citterio A, Arnoldi A, Panzeri E, Merlini L, D’Angelo MG, Musumeci O, Toscano T, Bondi A, Martinuzzi A, Bresolin N, Bassi MT (2015) Variants in KIF1A gene in dominant and sporadic forms of hereditary spastic paraparesis. J Neurol 262:2684–2690. https://doi.org/10.1007/s00415-015-7899-9
Volk A, Conboy E, Wical B, Patterson M, Kirmani S (2015) Whole-exome sequencing in the clinic: Lessons from six consecutive cases from the clinician’s perspective. Mol Syndromol 6:23–31. https://doi.org/10.1159/000371598
Megahed H, Nicouleau M, Barcia G, Medina-Cano D, Siquier-Pernet K, Bole-Feysot C, Parisot M, Masson C, Nitschké P, Rio M, Bahi-Buisson N, Desguerre I, Munnich A, Boddaert N, Colleaux L, Cantagrel V (2016) Utility of whole exome sequencing for the early diagnosis of pediatric-onset cerebellar atrophy associated with developmental delay in an inbred population. Orphanet J Rare Dis 11:57. https://doi.org/10.1186/s13023-016-0436-9
Hotchkiss L, Donkervoort S, Leach ME, Mohassel P, Bharucha-Goebel DX, Bradley N, Nguyen D, Hu Y, Gurgel-Giannetti J, Bönnemann CG (2016) Novel de novo mutations in KIF1A as a cause of hereditary spastic paraplegia with progressive central nervous system involvement. J Child Neurol 31:1114–1119. https://doi.org/10.1177/0883073816639718
Langlois S, Tarailo-Graovac M, Sayson B, Drögemöller B, Swenerton A, Ross CJD, Wasserman WW, Van Karnebeek CDM (2016) De novo dominant variants affecting the motor domain of KIF1A are a cause of PEHO syndrome. Eur J Hum Genet 24:949–953. https://doi.org/10.1038/ejhg.2015.217
Tomaselli PJ, Rossor AM, Horga A, Laura M, Blake JC, Houlden H, Reilly MM (2017) A de novo dominant mutation in KIF1A associated with axonal neuropathy, spasticity and autism spectrum disorder. J Peripher Nerv Syst 2:460–463. https://doi.org/10.1111/jns.12235
Raffa L, Matton MP, Michaud J, Rossignol E, Decarie JC, Ospina LH (2017) Optic nerve hypoplasia in a patient with a de novo KIF1A heterozygous mutation. Can J Ophthalmol 52:e169–e171. https://doi.org/10.1016/j.jcjo.2017.02.021
Travaglini L, Aiello C, Stregapede F, D’Amico A, Alesi V, Ciolfi A, Bruselles A, Catteruccia M, Pizzi S, Zanni G, Loddo S, Barresi S, Vasco G, Tartaglia M, Bertini E, Nicita F (2018) The impact of next-generation sequencing on the diagnosis of pediatric-onset hereditary spastic paraplegias: new genotype-phenotype correlations for rare HSP-related genes. Neurogenetics 19:111–121. https://doi.org/10.1007/s10048-018-0545-9
Wang J, Zhang Q, Chen Y, Yu S, Wu X, Bao X (2019) Rett and Rett-like syndrome: expanding the genetic spectrum to KIF1A and GRIN1 gene. Mol Genet Genom Med 7:1–8. https://doi.org/10.1002/mgg3.968
Pennings M, Schouten MI, van Gaalen J, Meijer RPP, de Bot ST, Kriek M, Saris CGJ, van den Berg LH, van Es MA, Zuidgeest DMH, Elting MW, van de Kamp JM, van Spaendonck-Zwarts KY, de Die-Smulders C, Brilstra EH, Verschuuren CC, de Vries BBA, Bruijn J, Sofou K, Duijkers FA, Jaeger B, Schieving JH, van de Warrenburg BP, Kamsteeg EJ (2020) KIF1A variants are a frequent cause of autosomal dominant hereditary spastic paraplegia. Eur J Hum Genet 28:40–49. https://doi.org/10.1038/s41431-019-0497-z
Spagnoli C, Rizzi S, Salerno GG, Frattini D, Fusco C (2019) Long-term follow-up until early adulthood in autosomal dominant, complex SPG30 with a novel KIF1A variant: a case report. Ital J Pediatr 45:155. https://doi.org/10.1186/s13052-019-0752-5
Van Eyk CL, Corbett MA, Frank MSB, Webber DL, Newman M, Berry JG, Harper K, Haines BP, McMichael G, Woenig JA, MacLennan AH, Gecz J (2019) Targeted resequencing identifies genes with recurrent variation in cerebral palsy. NPJ Genom Med 4:27. https://doi.org/10.1038/s41525-019-0101-z
Urtiaga Valle S, Fournier Gil B, Ramiro León MS, Martínez Menéndez B (2020) Usefulness of exome sequencing in the study of spastic paraparesis and cerebellar atrophy: de novo mutation of the KIF1A gene, a new hope in prognosis. Neurologia 35:535–538. https://doi.org/10.1016/j.nrl.2018.07.001
Nemani T, Steel D, Kaliakatsos M, DeVile C, Ververi A, Scott R, Getov S, Sudhakar S, Male A, Mankad K, Muntoni F, Reilly MM, Kurian MA, Carr L, Munot P (2020) KIF1A-related disorders in children: a wide spectrum of central and peripheral nervous system involvement. J Peripher Nerv Syst 25:117–124. https://doi.org/10.1111/jns.12368
Kurihara M, Ishiura H, Bannai T, Mitsui J, Yoshimura J, Morishita S, Hayashi T, Shimizu J, Toda T, Tsuji S (2020) A novel de novo KIF1A mutation in a patient with autism, hyperactivity, epilepsy, sensory disturbance, and spastic paraplegia. Intern Med 59:839–842. https://doi.org/10.2169/internalmedicine.3652-19
Van Beusichem ME, Nicolai J, Verhoeven J, Speth L, Coenen M, Willemsen MA, Kamsteeg EJ, Stumpel C, Vermeulen RJ (2020) Mobility characteristics of children with spastic paraplegia due to a mutation in the KIF1A gene. Neuropediatrics 51:146–153. https://doi.org/10.1055/s-0039-3400988
Kaur S, Van Bergen NJ, Verhey KJ, Nowell CJ, Budaitis B, Yue Y, Ellaway C, Brunetti-Pierri N, Cappuccio G, Bruno I, Boyle L, Nigro V, Torella A, Roscioli T, Cowley MJ, Massey S, Sonawane R, Burton MD, Schonewolf-Greulich B, Tümer Z, Chung WK, Gold WA, Christodoulou J (2020) Expansion of the phenotypic spectrum of de novo missense variants in kinesin family member 1A (KIF1A). Hum Mutat 41:1761–1774. https://doi.org/10.1002/humu.24079
Nicita F, Ginevrino M, Travaglini L, D’Arrigo S, Zorzi G, Borgatti R, Terrone G, Catteruccia M, Vasco G, Brankovic V, Siliquini S, Romano S, Veredice C, Pedemonte M, Armando M, Lettori D, Stregapede F, Bosco L, Sferra A, Tessarollo V, Romaniello R, Ristori G, Bertini E, Valente EM, Zanni G (2020) Heterozygous KIF1A variants underlie a wide spectrum of neurodevelopmental and neurodegenerative disorders. J Med Genet. https://doi.org/10.1136/jmedgenet-2020-107007
Rudenskaya GE, Kadnikova VA, Ryzhkova OP, Bessonova LA, Dadali EL, Guseva DS, Markova TV, Khmelkova DN, Polyakov AV (2020) KIF1A-related autosomal dominant spastic paraplegias (SPG30) in Russian families. BMC Neurol 20:1–14. https://doi.org/10.1186/s12883-020-01872-4
Cheon CK, Lim SH, Kim YM, Kim D, Lee NY, Yoon TS, Kim NS, Kim E, Lee JR (2017) Autosomal dominant transmission of complicated hereditary spastic paraplegia due to a dominant negative mutation of KIF1A, SPG30 gene. Sci Rep 7:12527. https://doi.org/10.1038/s41598-017-12999-9
Guo Y, Chen Y, Yang M, Xu X, Lin Z, Ma J, Chen H, Hu Y, Ma Y, Wang X, Tian X (2020) A rare KIF1A missense mutation enhances synaptic function and increases seizure activity. Front Genet 11:61. https://doi.org/10.3389/fgene.2020.00061
Ylikallio E, Kim D, Isohanni P, Auranen M, Kim E, Lönnqvist T, Tyynismaa H (2015) Dominant transmission of de novo KIF1A motor domain variant underlying pure spastic paraplegia. Eur J Hum Genet 23:1427–1430. https://doi.org/10.1038/ejhg.2014.297
Roda RH, Schindler AB, Blackstone C (2017) Multigeneration family with dominant SPG30 hereditary spastic paraplegia. Ann Clin Transl Neurol 4:821–824. https://doi.org/10.1002/acn3.452
Lu C, Li LX, Dong HL, Wei Q, Liu ZJ, Ni W, Gitler AD, Wu ZY (2018) Targeted next-generation sequencing improves diagnosis of hereditary spastic paraplegia in Chinese patients. J Mol Med 96:701–712. https://doi.org/10.1007/s00109-018-1655-4
González EM, Perona CL, González CG, De Castro CP, Pereira NG, Martín RDS, Muriel MAP, Gonda ES (2019) A novel heterozygous variant in the KIF1A gene causes a complicated form of spastic paraplegia. Clin Chim Acta 493:S243–S244. https://doi.org/10.1016/j.cca.2019.03.503
Carrasco Salas P, Martínez Fernández E, Méndez del Barrio C, Serrano Mira A, Guerrero Moreno N, Royo I, Delgado M, Oropesa J, Vázquez Rico I (2020) Clinical and molecular characterization of hereditary spastic paraplegia in a Spanish southern region. Int J Neurosci 15:1–12. https://doi.org/10.1080/00207454.2020.1838514
Iqbal Z, Rydning SL, Wedding IM, Koht J, Pihlstrøm L, Rengmark AH, Henriksen SP, Tallaksen CME, Toft M (2017) Correction: targeted high throughput sequencing in hereditary ataxia and spastic paraplegia. PLoS ONE 12(10):e0186571. https://doi.org/10.1371/journal.pone.0186571
Muir AM, Myers CT, Nguyen NT, Saykally J, Craiu D, De Jonghe P, Helbig I, Hoffman-Zacharska D, Guerrini R, Lehesjoki AE, Marini C, Møller RS, Serratosa J, Štěrbová K, Striano P, von Spiczak S, Weckhuysen S, Mefford HC (2019) Genetic heterogeneity in infantile spasms. Epilepsy Res 156:106181. https://doi.org/10.1016/j.eplepsyres.2019.106181
Kashimada A, Hasegawa S, Nomura T, Shiraku H, Moriyama K, Suzuki T, Nakajima K, Mizuno T, Imai K, Sugawara Y, Morio T, Kumada S, Takagi M (2019) Genetic analysis of undiagnosed ataxia-telangiectasia-like disorders. Brain Dev 41:150–157. https://doi.org/10.1016/j.braindev.2018.09.007
Taşkıran EZ, Karaosmanoğlu B, Koşukcu C, Ürel-Demir G, Akgün-Doğan Ö, Şimşek-Kiper PÖ, Alikaşifoğlu M, Boduroğlu K, Utine GE (2021) Diagnostic yield of whole-exome sequencing in non-syndromic intellectual disability. J Intellect Disabil Res 65:577–588. https://doi.org/10.1111/jir.12835
Montenegro-Garreaud X, Hansen AW, Khayat MM, Chander V, Grochowski CM, Jiang Y, Li H, Mitani T, Kessler E, Jayaseelan J, Shen H, Gezdirici A, Pehlivan D, Meng Q, Rosenfeld JA, Jhangiani SN, Madan-Khetarpal S, Scott DA, Abarca-Barriga H, Trubnykova M, Gingras MC, Muzny DM, Posey JE, Liu P, Lupski JR, Gibbs RA (2020) Phenotypic expansion in KIF1A-related dominant disorders: a description of novel variants and review of published cases. Hum Mutat 41:2094–2104. https://doi.org/10.1002/humu.24118
Klebe S, Lossos A, Azzedine H, Mundwiller E, Sheffer R, Gaussen M, Marelli C, Nawara M, Carpentier W, Meyer V, Rastetter A, Martin E, Bouteiller D, Orlando L, Gyapay G, El-Hachimi KH, Zimmerman B, Gamliel M, Misk A, Lerer I, Brice A, Durr A, Stevanin G (2012) KIF1A missense mutations in SPG30, an autosomal recessive spastic paraplegia: distinct phenotypes according to the nature of the mutations. Eur J Hum Genet 20:645–649. https://doi.org/10.1038/ejhg.2011.261
Erlich Y, Edvardson S, Hodges E, Zenvirt S, Thekkat P, Shaag A, Dor T, Hannon GJ, Elpeleg O (2011) Exome sequencing and disease-network analysis of a single family implicate a mutation in KIF1A in hereditary spastic paraparesis. Genome Res 21:658–664. https://doi.org/10.1101/gr.117143.110
Krenn M, Zulehner G, Hotzy C, Rath J, Stogmann E, Wagner M, Haack TB, Strom TM, Zimprich A, Zimprich F (2017) Hereditary spastic paraplegia caused by compound heterozygous mutations outside the motor domain of the KIF1A gene. Eur J Neurol 24:741–747. https://doi.org/10.1111/ene.13279
Hosokawa S, Kubo Y, Arakawa R, Takashima H, Saito K (2020) Analysis of spinal muscular atrophy-like patients by targeted resequencing. Brain Dev 42:148–156. https://doi.org/10.1016/j.braindev.2019.10.008
Dosi C, Pasquariello R, Ticci C, Astrea G, Trovato R, Rubegni A, Tessa A, Cioni G, Santorelli FM, Battini R (2021) Neuroimaging patterns in paediatric onset hereditary spastic paraplegias. J Neurol Sci 425:117441. https://doi.org/10.1016/j.jns.2021.117441
Synofzik M, Schüle R (2017) Overcoming the divide between ataxias and spastic paraplegias: shared phenotypes, genes, and pathways. Mov Disord 32:332–345. https://doi.org/10.1002/mds.26944
Schüle R, Holland-Letz T, Klimpe S, Kassubek J, Klopstock T, Mall V, Otto S, Winner B, Schöls L (2006) The Spastic Paraplegia Rating Scale (SPRS): a reliable and valid measure of disease severity. Neurology 67:430–434. https://doi.org/10.1212/01.wnl.0000228242.53336.90
Fink JK (2013) Hereditary spastic paraplegia: clinico-pathologic features and emerging molecular mechanisms. Acta Neuropathol 126:307–328. https://doi.org/10.1007/s00401-013-1115-8
Li M, Zheng W (2011) Probing the structural and energetic basis of kinesin-microtubule binding using computational alanine-scanning mutagenesis. Biochemistry 50:8645–8655. https://doi.org/10.1021/bi2008257
Kalantari S, Filges I (2020) “Kinesinopathies”: emerging role of the kinesin family member genes in birth defects. J Med Genet 57:797–807. https://doi.org/10.1136/jmedgenet-2019-106769
Wang D, Nitta R, Morikawa M, Yajima H, Inoue S, Shigematsu H, Kikkawa M, Hirokawa N (2016) Motility and microtubule depolymerization mechanisms of the Kinesin-8 motor, KIF19A. Elife 5:e18101. https://doi.org/10.7554/eLife.18101
Luo T, Li K, Ling Z, Zhao G, Li B, Wang Z, Wang X, Han Y, Xia L, Zhang Y, Zhou Q, Fang Z, Wang Y, Chen Q, Zhou X, Pan H, Zhao Y, Wang Y, Dong L, Huang Y, Hu Z, Pan Q, Xia K, Li J (2021) De novo mutations in folate-related genes associated with common developmental disorders. Comput Struct Biotechnol J 19:1414–1422. https://doi.org/10.1016/j.csbj.2021.02.011
Boyle WKCL, Rao L, Kaur S, Fan X, Mebane C, Hamm L, Thornton A, Ahrendsen JT, Anderson MP, Christodoulou J, Gennerich A, Shen Y, Chung WK (2021) Genotype and defects in microtubule-based motility correlate with clinical severity in KIF1A-associated neurological disorder. HGG Adv. 2(2):100026. https://doi.org/10.1016/j.xhgg.2021.100026
Tarabeux J, Champagne N, Brustein E, Hamdan FF, Gauthier J, Lapointe M, Maios C, Piton A, Spiegelman D, Henrion É, Millet B, Rapoport JL, Delisi LE, Joober R, Fathalli F, Fombonne É, Mottron L, Forget-Dubois N, Boivin M, Michaud JL, Lafrenire RG, Drapeau P, Krebs MO, Ouleau GA (2010) De novo truncating mutation in kinesin 17 associated with schizophrenia. Biol Psychiatry 68:649–656. https://doi.org/10.1016/j.biopsych.2010.04.018
Afenjar A, Billette de Villemeur T, Mignot C, Guet A, Keren B, Valence S, Garel C, Burglen L (2017) De novo mutations in KIF1A in 2 patients with congenital ataxia. In: Abstracts of EPNS 2017—12th European Paediatric Neurology Society Congress: Lyon, France20–24 June 2017. Eur J Ped Neurol 21:e1–254
Lo RY, Figueroa KP, Pulst SM, Lin CY, Perlman S, Wilmot G, Gomez G, Schmahmann J, Paulson H, Shakkottai VG, Ying S, Zesiewicz T, Bushara K, Geschwind M, Xia G, Subramony SH, Ashizawa T, Kuo SH (2015) Coenzyme Q10 and spinocerebellar ataxias. Mov Disord 30:214–220. https://doi.org/10.1002/mds.26088
Pyle A, Nightingale HJ, Griffin H, Abicht A, Kirschner J, Baric I, Cuk M, Douroudis K, Feder L, Kratz M, Czermin B, Kleinle S, Santibanez-Koref M, Karcagi V, Holinski-Feder E, Chinnery PF, Horvath R (2015) Respiratory chain deficiency in nonmitochondrial disease. Neurol Genet 1:e6. https://doi.org/10.1212/NXG.0000000000000006
Lo Giudice T, Lombardi F, Santorelli FM, Kawarai T, Orlacchio A (2014) Hereditary spastic paraplegia: clinical-genetic characteristics and evolving molecular mechanisms. Exp Neurol 261:518–539. https://doi.org/10.1016/j.expneurol.2014.06.011
Acknowledgements
We thank Catherine J. Wrenn for expert editing of the manuscript and critical advice on tables and figures. We are indebted to our colleagues who referred patients with spastic-ataxia for gene screening.
Funding
This research was partially supported by Grants MITO-NEXT and MINDFUL CC-2019-2366613 (to FMS), RC 5 × 1000 2020–2021 (to AT, RB, and AR), and RF-2019-12370112 (to AT and MTB) from the Italian Ministry of Health. We also acknowledge the financial support of DECODE-EE Tuscany Region Call for Health 2018 (to RG).
Author information
Authors and Affiliations
Contributions
Conceptualization: [SDV, AT, AR, CD, FMS]; Acquisition data: [SDV, AT, AR, CD, AA, RP, GA, MTB, RB, CC, EC, GC, GDM, ARF, AF, CF, CF, SG, CG, RG, SH, DL, FM, AMa, AMi, AP, AMP, FP, GP, EP, RR, AR, CSan, SS, CSp, CT]. Methodology: [SDV, AT, CD, AR]; Formal analysis and investigation: [SDV, AT, CD, AR, JB, RP]; Writing—original draft preparation: [SDV, AT, CD, AR, JB, FMS]; Writing—review and editing: [SDV, AT, AR, FMS]; Critical revision: [SDV, AT, AR, CD, RP, GA, MTB, RB, CC, EC, GC, GDM, ARF, AF, CF, CF, SG, RG, EPa, DL, FM, AM, AP, AMP, FP, GP, EPr, AR, SS, CS, CT, FMS]; Funding acquisition: [FMS]; Resources: [FMS, RG]; Supervision: [FMS, AT]. All authors fully comply with and approve the version to be published.
Corresponding authors
Ethics declarations
Conflicts of interest
The authors declare no conflict of interest.
Ethical standards
This study was approved by the Tuscany Regional Pediatric Ethics Committee and written informed consent was obtained from the patients.
Supplementary Information
Below is the link to the electronic supplementary material.
Appendix
. Detailed genetic information on KIF1A variants reported in our cohort. (XLSX 13 KB)
Figure S1
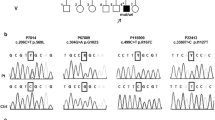
. Family pedigrees. Figure S2. (A) Pie chart and histogram showing the clinical manifestations in our cohort of 28 patients. The relative Human Phenotype Ontology codes are given in brackets. (B) Histogram showing the neuroradiological findings (MRI) in 27 members of our cohort. The relative Human Phenotype Ontology codes are given in brackets. Figure S3. (A) Histogram showing the clinical manifestations of the patients with congenital ataxia in our cohort (n=6). The relative Human Phenotype Ontology codes are given in brackets. GDD: Global developmental delay. ID: Intellectual disability. (B) Histogram showing the neuroradiological findings in the patients with congenital ataxia in our cohort (n=6). The relative Human Phenotype Ontology codes are given in brackets. Figure S4. Myo-pathological changes in patients with KIF1A mutations: a) Hematoxylin and eosin staining demonstrating marked variation in fiber size in Pt9. b) Oil Red O Stain showing mild lipid storage in Pt5. (PDF 1167 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Vecchia, S.D., Tessa, A., Dosi, C. et al. Monoallelic KIF1A-related disorders: a multicenter cross sectional study and systematic literature review. J Neurol 269, 437–450 (2022). https://doi.org/10.1007/s00415-021-10792-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00415-021-10792-3