Abstract
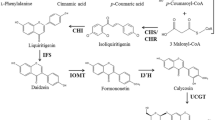
Fruits of Actinidia arguta (Sieb. et Zucc.) Planch. ex Miq. are important source of bioactive substances including flavonoids. However, there were few reports on the phytochemical aspects of Actinidia arguta in different tissues. Here, we investigated the flavonoid content in different tissues of A. arguta by LC–MS/MS technology. In our study, ten flavonoids were detected, namely kaempferol-3-O-rutinoside (+), kaempferol-3-O-rutinoside (−), kaempferol-3-O-neohesperidoside (P), isorhamnetin-3-O-α-l-rhamnopyranosyl-(1→3)-α-l-rhamnopyranosyl-(1→ 6)-β-d-galactopyranoside, isorhamnetin-3-O-α-l-rhamnopyranosyl-(1→3)-α-l-rhamnopyranosyl-(1→6)-β-d-galactopyranoside, isorhamnetin-3-O-neohesperidoside (P)(+), isorhamnetin-3-O-neohesperidoside (P)(−), isorhamnetin-3-O-rutinoside, isorhamnetin-3-O-neohespeidoside, quercetin -3-O- rhamnoglycoside, which were belonged to kaempferol, isorhamnetin and quercetin, the major flavonoid compounds in Actinidia arguta, which decreased with the order of leaf > petiole > fruit. In young apical leaves, the total flavonoid content was 6.3-fold higher than that in fruit. And there was a continuous decrease as leaf development in total flavonoids content. In addition, except AaCHS-2 and AaDFR, the expression levels of nine genes-related flavonoid biosynthesis were similar to the change of total flavonoids content in different tissues. It indicated that the eleven genes played main regulation in flavonoid biosynthesis. Thus, our results revealed the regulatory information of flavonoid biosynthesis in Actinidia arguta leaves and provide technical support for the full utilization of leaves.





Similar content being viewed by others
References
Ajila C, Naidu K, Bhat S, Rao U (2007) Bioactive compounds and antioxidant potential of mango peel extract. Food Chem 105:982–988. https://doi.org/10.1016/j.foodchem.2007.04.052
Almeida D, Pinto D, Santos J, Vinha AF, Palmeira J, Ferreira HN, Rodrigues F, Oliveira MBPP (2018) Hardy kiwifruit leaves (Actinidia arguta): an extraordinary source of value-added compounds for food industry. Food Chem 259:113–121. https://doi.org/10.1016/j.foodchem.2018.03.113
Almeida MMB, de Sousa PHM, Arriaga ÂMC, do Prado GM, de Carvalho Magalhães CE, Maia GA, de Lemos TLG (2011) Bioactive compounds and antioxidant activity of fresh exotic fruits from northeastern Brazil. Food Res Int 44:2155–2159. https://doi.org/10.1016/j.foodres.2011.03.051
Arulmozhi S, Mazumder PM, Sathiyanarayanan L, Ashok P (2011) Anti-arthritic and antioxidant activity of leaves of Alstonia scholaris Linn. R.Br. Eur J Integr Med 3:e83–e90. https://doi.org/10.1016/j.eujim.2011.04.019
Cho Y, Cho H, Park M (2011) The distribution, characteristics and utilization of Korean native Actinidia genus. Korean J Org Agric 19:30–34
Delgado-Adámez J, Fernández-León MF, Velardo-Micharet B, González-Gómez D (2012) In vitro assays of the antibacterial and antioxidant activity of aqueous leaf extracts from different Prunus salicina Lindl. cultivars. Food Chem Toxicol 50:2481–2486. https://doi.org/10.1016/j.fct.2012.02.024
Dizdarevic LL, Biswas D, Uddin MD. M, Jørgenesen A, Falch E, Bastani NE, Duttaroy AK (2014) Inhibitory effects of kiwifruit extract on human platelet aggregation and plasma angiotensin-converting enzyme activity. Platelets 25(8):567–575. https://doi.org/10.3109/09537104.2013.852658
Du QH, Zhang QY, Han T, Jiang YP, Peng C, Xin HL (2016) Dynamic changes of flavonoids in Actinidia valvata leaves at different growing stages measured by HPLC-MS/MS. Chin J Nat Med 14:66–72. https://doi.org/10.3724/SP.J.1009.2015.00066
Ferguson AR, Huang H (2007) Genetic resources of kiwifruit: domestication and breeding. Hortic Rev. https://doi.org/10.1002/9780470168011.ch1
Gupta N, Sharma SK, Rana JC, Chauhan RS (2011) Expression of flavonoid biosynthesis genes vis-à-vis rutin content variation in different growth stages of Fagopyrum species. J Plant Physiol 168:2117–2123. https://doi.org/10.1016/j.jplph.2011.06.018
Han M, Yang C, Zhou J, Zhu J, Meng J, Shen T, Xin Z, Li H (2020) Analysis of flavonoids and anthocyanin biosynthesis-related genes expression reveals the mechanism of petal color fading of Malus hupehensis (Rosaceae). Braz J Bot. https://doi.org/10.1007/s40415-020-00590-y
Jang DS, Lee GY, Lee YM, Kim YS, Sun H, Kim DH, Kim JS (2009) Flavan-3-ols having a γ-Lactam from the roots of Actinidia arguta inhibit the formation of advanced glycation end products in vitro. Chem Pharm Bull 57:397–400. https://doi.org/10.1248/cpb.57.397
Jin X, Huang H, Wang L, Sun Y, Dai SL (2016) Transcriptomics and metabolite analysis reveals the molecular mechanism of anthocyanin biosynthesis branch pathway in different Senecio cruentus cultivars. Front Plant Sci. https://doi.org/10.3389/fpls.2016.01307
Kabaluk JT, Kempler C, Toivonen PMA (1997) Actinidia arguta: characteristics relevant to commercial production. Fruit Varieties J 51:117–122. https://doi.org/10.1007/s001220050500
Khoobi M, Emami S, Dehghan G, Foroumadi A, Ramazani A, Shafiee A (2011) Synthesis and free radical scavenging activity of coumarin derivatives containing a 2-methylbenzothiazoline motif. Arch Pharm 344:588–594. https://doi.org/10.1002/ardp.201000271
Leontowicz H, Leontowicz M, Latocha P, Jesion I, Park YS, Katrich E, Barasch D, Nemirovski A, Gorinstein S (2016) Bioactivity and nutritional properties of hardy kiwifruit Actinidia arguta in comparison with Actinidia deliciosa ‘Hayward’ and Actinidia eriantha ‘Bidan.’ Food Chem 196:281–291. https://doi.org/10.1016/j.foodchem.2015.08.127
Li W, Liu Y, Zeng S, Xiao G, Wang G, Wang Y, Peng M, Huang H (2015) Gene expression profiling of development and anthocyanin accumulation in kiwifruit (Actinidia chinensis) based on transcriptome sequencing. PLoS ONE 10:e0136439. https://doi.org/10.1371/journal.pone.0136439
Li WH, Feng JR, Zhang SK, Tang ZH (2019) The correlation between cellular features and gene expression in “Korla” fragrant pear. HortScience 55:1–6. https://doi.org/10.21273/HORTSCI14613-19
Li Y, Fang J, Qi X, Lin M, Zhong Y, Sun L, Cui W (2018a) Combined analysis of the fruit metabolome and transcriptome reveals candidate genes involved in flavonoid biosynthesis in Actinidia arguta. Int J Mol Sci 19:1471. https://doi.org/10.3390/ijms19051471
Li Y, Fang J, Qi X, Lin M, Zhong Y, Sun L (2018b) A key structural gene, AaLDOX, is involved in anthocyanin biosynthesis in all red-fleshed kiwifruit (Actinidia arguta) based on transcriptome analysis. Gene 648:31–41. https://doi.org/10.1016/j.gene.2018.01.022
Li Y, Qi X, Cui W, Lin M, Qiao C, Zhong Y, Fang J, Hu C (2020) Restraint of bagging on fruit skin coloration in on-tree kiwifruit (Actinidia arguta). J Plant Growth Regul. https://doi.org/10.1007/s00344-020-10124-1
Liu Y, Zhou B, Qi Y, Chen X, Liu C, Liu Z, Ren X (2017) Expression differences of pigment structural genes and transcription factors explain flesh coloration in three contrasting kiwifruit cultivars. Front Plant Sci. https://doi.org/10.3389/fpls.2017.01507
Matich AJ, Young H, Allen JM, Wang MY, Fielder S, McNeilage MA, MacRae EA (2003) Actinidia arguta: volatile compounds in fruit and flowers. Phytochemistry 63:285–301. https://doi.org/10.1016/s0031-9422(03)00142-0
M’mbone ME, Cheng W, Xu L, Wang Y, Karanja BK, Zhu X, Cao Y, Liu L (2018) Identification and transcript analysis of MATE genes involved in anthocyanin transport in radish (Raphanus sativus L.). Sci Hortic-Amst 238:195–203. https://doi.org/10.1016/j.scienta.2018.04.029
Moriguchi T, Kita M, Tomono Y, Endo-Inagaki T, Omura M (2001) Gene expression in flavonoid biosynthesis: correlation with flavonoid accumulation in developing citrus fruit. Physiol Plantarum 111:66–74. https://doi.org/10.1034/j.1399-3054.2001.1110109.x
Nakatsuka A, Mizuta D, Kii Y, Miyajima I, Kobayashi N (2008) Isolation and expression analysis of flavonoid biosynthesis genes in evergreen azalea. Sci Hortic-Amst 118:314–320. https://doi.org/10.1016/j.scienta.2008.06.016
Nile SH, Keum YS, Nile AS, Jalde SS, Patel RV (2017) Antioxidant, anti-inflammatory, and enzyme inhibitory activity of natural plant flavonoids and their synthesized derivatives. J Biochem Mol Toxic 32:e22002. https://doi.org/10.1002/jbt.22002
Park D, Park Y, Lee YH, Choi IY, Park KC, Park SU, Sang UP, Kim BS, Yeoung YR, Park NI (2017) A comparative study of phenolic antioxidant activity and flavonoid biosynthesis-related gene expression between summer and winter strawberry cultivars. J Food Sci 82:341–349. https://doi.org/10.1111/1750-3841.13600
Payyavula RS, Navarre DA, Kuhl J, Pantoja A (2013) Developmental effects on phenolic, flavonol, anthocyanin, and carotenoid metabolites and gene expression in potatoes. J Agric Food Chem 61:7357–7365. https://doi.org/10.1021/jf401522k
Peng Y, Lin-Wang K, Cooney JM, Wang T, Espley RV, Allan AC (2019) Differential regulation of the anthocyanin profile in purple kiwifruit (Actinidia species). Hortic Res. https://doi.org/10.1038/s41438-018-0076-4
Singh AP, Wilson T, Luthria D, Freeman MR, Scott RM, Bilenker D, Shah S, Somasundaram S, Vorsa N (2011) LC-MS–MS characterisation of curry leaf flavonols and antioxidant activity. Food Chem 127:80–85. https://doi.org/10.1016/j.foodchem.2010.12.091
Tan C, Wang Z, Feng X, Irfan M, Liu C (2021) Identification of bioactive compounds in leaves and fruits of Actinidia arguta accessions from northeastern China and assessment of their antioxidant activity with a radical-scavenging effect. Biotechnol Biotechnol Equip 35:593–607. https://doi.org/10.1080/13102818.2021.1908166
Webby RF (1991) A flavonol triglycoside from Actinidia arguta var Giraldii. Phytochemistry 30:2443–2444. https://doi.org/10.1016/0031-9422(91)83680-j
Wojdylo A, Nowicka P (2019) Anticholinergic effects of Actinidia arguta fruits and their polyphenol content determined by liquid chromatography-photodiode array detector-quadrupole/time off light-mass spectrometry (LC-MS-PDA-Q/TOF). Food Chem 271:216–223. https://doi.org/10.1016/j.foodchem.2018.07.084
Xu B, Wang M, Cao J, Wang Y (2013) Differences in antioxidant components and activity of pear jujube (Zizyphus jujube Mill. Cv. Lizao) leaf tea among different growth stages. Food Sci. https://doi.org/10.7506/spkx1002-6630-201313008 (in Chinese)
Xue L, Wang Z, Zhang W, Li Y, Wang J, Lei J (2016) Flower pigment heritance and anthocyanin characterization of hybrids from pink-flowered and white-flowered strawberry. Sci Hortic-Amst 200:143–150. https://doi.org/10.1016/j.scienta.2016.01.020
Yu M, Man Y, Lei R, Lu X, Wang Y (2020) Metabolomics study of flavonoids and anthocyanin-related gene analysis in kiwifruit (Actinidia chinensis) and kiwiberry (Actinidia arguta). Plant Mol Biol Rep. https://doi.org/10.1007/s11105-020-01200-7
Yuan D, Chen X, Hou L, Jiang Q, Liu F, Zhu X (2016) Study on production of Ginkgo biloba tea with pile-fermentation. J Food Sc Biotechnol 36:84–89 (in Chinese)
Zainudina MAM, Hamida AA, Anwar F, Osman A, Saari N (2014) Variation of bioactive compounds and antioxidant activity of carambola (Averrhoa carambola L.) fruit at different ripening stages. Sci Hortic-Amst 172:325–331. https://doi.org/10.1016/j.scienta.2014.04.007
Zhang FS, Wang QY, Pu YJ, Chen TY, Qin XM, Gao J (2017) Identification of genes involved in flavonoid biosynthesis in Sophora japonica through transcriptome sequencing. Chem Biodivers 14:e1700369. https://doi.org/10.1002/cbdv.201700369
Zhang L, Gao Y, Zhang Y, Liu J, Yu J (2010) Changes in bioactive compounds and antioxidant activities in pomegranate leaves. Sci Hortic-Amst 123:543–546. https://doi.org/10.1016/j.scienta.2009.11.008
Zhao X, Guo S, Yan H, Lu Y, Zhang F, Qian D, Wang H, Duan J (2019) Analysis of phenolic acids and flavonoids in leaves of Lycium barbarum from different habitats by ultra-high-performance liquid chromatography coupled with triple quadrupole tandem mass spectrometry. Biomed Chromatogr. https://doi.org/10.1002/bmc.4552
Zhou Z, Cao Y, Wang Y, Cheng D, Mi J, Li X, Yan Y, Zeng X (2017) Analysis of chemical compositions and antioxidant activities of Lycium barbarum bud and leaf teas. Sci Technol Food Ind 38:129–145 (in Chinese)
Zuo LL, Wang ZY, Fan ZL, Tian SQ, Liu JR (2012) Evaluation of antioxidant and antiproliferative properties of three Actinidia (Actinidia kolomikta, Actinidia arguta, Actinidia chinensis) extracts in vitro. Int J Mol Sci 13:5506–5518. https://doi.org/10.3390/ijms13055506
Zupkó I, Hohmann J, Rédei D, Falkay G, Janicsák G, Máthé I (2001) Antioxidant activity of leaves of Salvia species in enzyme-dependent and enzyme-independent systems of lipid peroxidation and their phenolic constituents. Planta Med 67:366–368. https://doi.org/10.1055/s-2001-14327
Acknowledgements
The authors thank Prof. Lei Jiajun's team and Associate Prof. Dai Hanping’s team for providing language and writing assistance.
Funding
The authors declare the following financial interests/personal relationships which may be considered as potential competing interests:
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Tan, C., Wang, Z., Irfan, M. et al. Analysis of flavonoids biosynthesis-related genes expression reveals the mechanism of difference of flavonoid content in different tissues of Actinidia arguta. Braz. J. Bot 44, 513–523 (2021). https://doi.org/10.1007/s40415-021-00737-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40415-021-00737-5