Abstract
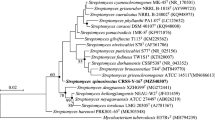
An aerobic, non-motile, Gram-stain positive actinomycete, designated strain CA3R110T, was isolated from the surface-sterilised root of Coffea arabica L. collected from Lampang Province, Thailand. 16S rRNA gene sequence analysis indicated that strain CA3R110T was a member of the genus Streptomyces and showed the closest similarities to Streptomyces buecherae AC541T (99.2%), followed by Streptomyces rapamycinicus NRRL B-5491T (99.1%), Streptomyces luteoverticillatus NBRC 3840T (99.1%), Streptomyces coerulescens NBRC 12758T (99.1%), and Streptomyces iranensis HM 35T (99.0%). Strain CA3R110T contained LL-diaminopimelic acid in cell peptidoglycan, MK-9(H6), and MK-9(H8) as major menaquinone, iso-C16:0, iso-C15:0, C16:0 as major fatty acids. Diphosphatidylglycerol, phosphatidylethanolamine, hydroxy-phosphatidylethanolamine, phosphatidylglycerol, phosphatidylinositol, phosphatidylinositolmannoside were detected in the cell. The chemotaxonomic characteristics possessed the typical properties of the genus Streptomyces. A low digital DNA–DNA hybridization (< 55.7%) and average nucleotide identity-blast (ANIb) (< 92.2%) values revealed that strain CA3R110T could be distinguished from any known Streptomyces species. With the differences in phenotypic and genotypic data, strain CA3R110T represents a novel species of genus Streptomyces, for which the name Streptomyces endocoffeicus sp. nov. is proposed. The type strain is CA3R110T (= TBRC 11245T = NBRC 114296T).

Similar content being viewed by others
Data availability
The DDBJ accession number for the 16S rRNA gene sequence of strain CA3R110T is MN116545. The whole-genome shotgun project has been deposited at DDBJ/ENA/GenBank under the accession JAERRG000000000. The version described in this paper is version JAERRG010000000.
Abbreviations
- ANIb:
-
Average nucleotide identity-BLAST
- ANIm:
-
Average nucleotide identity-MUMmer algorithm
- DAP:
-
Diaminopimelic acid
- dDDH:
-
Digital DNA-DNA hybridisation
- DPG:
-
Diphosphatidylglycerol
- PE:
-
Phosphatidylethanolamine
- OH-PE:
-
Hydroxy-phosphatidylethanolamine
- PG:
-
Phosphatidylglycerol
- PI:
-
Phosphatidylinositol
- PL:
-
Unidentified phospholipid
- NPG:
-
Ninhydrin-positive lipid
- L:
-
Unidentified lipid
- GGDC:
-
Genome to genome distance calculator
- HPLC:
-
High performance liquid chromatography
- ISP:
-
International Streptomyces Project
- ISCC-NBS:
-
Inter-Society Color Council-the National Bureau of Standards
- MK:
-
Menaquinone
- ML:
-
Maximum-likelihood
- MP:
-
Maximum-parsimony
- NJ:
-
Neighbour-joining
- PE:
-
Phosphatidylethanolamine
- PI:
-
Phosphatidylinositol
- TLC:
-
Thin layer chromatography
- TYGS:
-
Type Strain Genomic Server
References
AI-Tai A, Kim B, Kim SB, Manfiol GP, Goodfellow M, (1999) Streptomyces malaysiensis sp. nov., a new streptomycete species with rugose, ornamented spores. Int J Syst Bacteriol 49:1395–1402
Arai T (1975) Culture media for actinomycetes. The Society for Actinomycetes Japan, Tokyo
Aziz RK, Bartels D, Best AA, DeJongh M, Disz T, Edwards RA et al (2008) The RAST server: rapid annotations using subsystems technology. BMC Genom 8:9–75
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Com Biol 19:455–477
Cao P, Li C, Wang H, Yu Z, Xu X, Wang X, Zhao J, Xiang W (2020) Community structures and antifungal activity of root-associated endophytic actinobacteria in healthy and diseased cucumber plants and Streptomyces sp. HAAG3-15 as a promising biocontrol agent. Microor 8:236–253
Chun J, Oren A, Ventosa A, Christensen H, Arahal DR, Costa MS, Rooney AP, Yi H, Xu XW, Meyer SD, Trujillo ME (2018) Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol 68:461–466
Collins MD, Jones D (1980) Lipids in the classification and identification of coryneform bacteria containing peptidoglycans based on 2,4-diaminobutyric acid. J Appl Bacteriol 48:459–470
Collins MD, Pirouz T, Goodfellow M, Minnikin DE (1977) Distribution of menaquinones in actinomycetes and corynebacteria. J Gen Microbiol 100:221–230
Ezaki T, Hashimoto Y, Yabuuchi E (1989) Fluorometric deoxyribonucleic acid-deoxyribonucleic acid hybridization in microdilution wells as an alternative to membrane filter hybridization in which radioisotopes are used to determine genetic relatedness among bacterial strains. Int J Syst Bacteriol 39:224–229
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Gordon RE, Barnett DA, Handerhan JE, Pang CHN (1974) Nocardia coeliaca, Nocardia autotrophica, and the nocardin strain. Int J Syst Bacteriol 24:54–63
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hamm PS, Dunlap CA, Mullowney MW, Caimi NA, Kelleher NL, Thomson RJ, Alfaro AP, Northup DN (2020) Streptomyces buecherae sp. Nov., an actinomycete isolated from multiple bat species. Antonie Van Leeuwenhoek. https://doi.org/10.1007/s10482-020-01493-4
Hasegawa T, Takizawa M, Tanida S (1983) A rapid analysis for chemical grouping of aerobic actinomycetes. J Gen Appl Microbiol 29:319–322
Kaewkla O, Franco CMM (2021) Genome mining and description of Streptomyces albidus sp. nov., an endophytic actinobacterium with antibacterial potential. Antonie Van Leeuwenhoek. https://doi.org/10.1007/s10482-021-01539-1
Kämpfer P, Kroppenstedt RM (1996) Numerical analysis of fatty acid patterns of coryneform bacteria and related taxa. Can J Microbiol 42:989–1005
Kelly KL (1964) Inter-society color council: National Bureau Of Standard color name charts illustrated with centroid colors. US Government Printing Office, Washington DC
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Komagata K, Suzuki KI (1987) Lipid and cell-wall analysis in bacterial systematics. Methods Microbiol 19:161–207
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549
Labeda DP, Lyons AJ (1991) The Streptomyces violaceusniger cluster is heterogeneous in DNA relatedness among strains: emendation of the description of S violaceusniger and Streptomyces hygroscopicus. Int J Syst Bacteriol 41:398401
Meier-Kolthoff JP, Göker M (2019) TYGS is an automated highthroughput platform for state-of-the-art genome-based taxonomy. Nat Commun 10:2182
Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinf 14:60–73
Minnikin DE, O’Donnell AG, Goodfellow M, Alderson G, Athalye M, Schaala A, Parletta JH (1984) An integrated procedure for the extraction of bacterial isoprenoid quinones and polar lipids. J Microbiol Methods 2:233–241
Overbeek R, Olson R, Pusch GD, Olsen GJ, Davis JJ, Disz T et al (2014) The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res 42:D20–D214
Peng C, Zhuang X, Gao C, Wang Z, Zhao J, Huang SX, Liu C, Xiang W (2021) Streptomyces typhae sp. Nov., a novel endophytic actinomycete with antifungal activity isolated the root of cattail (Typha angustifolia L.). Anto van Leeu. https://doi.org/10.1007/s10482-021-01561-3
Pridham TG, Hesseltine CW, Benedict RG (1958) A guide for the classification of streptomycetes according to selected groups; placement of strains in morphological sections. Appl Microbiol 6:52–79
Richter M, Rosselló-Móra R (2009) Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA 106:19126–19131
Richter M, Rosselló-Móra R, Glöckner FO, Peplies J (2016) JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32:929–931
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sasaki T, Igarashi Y, Ogawa M, Furumai T (2002) Identification of 6-prenylindole as an antifungal metabolite of Streptomyces sp. TP-A0595 and synthesis and bioactivity of 6-substituted indoles. J Antibiot 55:1009–1012
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids. MIDI Technical Note 101. Microbial ID, Inc, Newark
Shirling EB, Gottlieb D (1966) Methods for characterization of Streptomyces species. Int J Syst Bacteriol 16:313–340
Shirling EB, Gottlieb D (1968) Cooperative Description of type cultures of Streptomyces III. Additional species descriptions from first and second studies. Int J Syst Bacteriol 18:279–392
Singh R, Dubey AK (2018) Diversity and applications of endophytic actinobacteria of plants in special and other ecological niches. Front Microbiol 9:1767
Solecka J, Zajko J, Postek M, Rajnisz A (2012) Biologically active secondary metabolites from Actinomycetes. Cent Eur J Biol 7:373–390
Tamaoka J (1994) Determination of DNA Base Composition. In: Goodfellow M, O’Donnell AG (eds) Chemical methods in prokaryotic systematics. Wiley, Chichester, pp 463–470
Tamaoka J, Katayama-Fujimura Y, Kuraishi H (1983) Analysis of bacterial menaquinone mixtures by high performance liquid chromatography. J Appl Bacteriol 54:31–36
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526
Thawai C (2015) Micromonospora costi sp. nov., isolated from a leaf of Costus speciosus. Int J Syst Evol Microbiol 55:1456–1461
Waksman SA (1961) The actinomycetes, classification, identification and description of genera and species, vol 2. Williams and Wilkins, Baltimore
Waksman SA, Henrici AT (1943) The nomenclature and classification of the actinomycetes. J Bacteriol 46:337–341
Wang Z, Tian J, Li X, Gan L, He L, Chu Y, Tian Y (2018) Streptomyces dioscori sp. nov., a novel endophytic actinobacterium isolated from bulbil of Dioscorea bulbifera L. Curr Microbiol 10:1384–1390
Wayne LG, Brenner DJ, Colwell RR, Grimont PAD, Kandler O, Krichevsky MI, Moore LH, Moore WEC, Murray RGE, Stackebrandt E, Starr MP, Truper HG (1987) Report of the ad hoc committee on reconciliation of approaches to bacterial systematics. Int J Syst Bacteriol 37:463–464
Wellington EMH, Stackebrandt E, Sanders D, Wolstrup J, Jorgensen NOG (1992) Taxonomic Status of Kitasatosporia, and Proposed Unification with Streptomyces on the Basis of Phenotypic and 16S rRNA Analysis and Emendation of Streptomyces Waksman and Henrici 1943, 339AL. Int J Syst Bact 42:156–160
Williams ST, Cross T (1971) Actinomycetes. In: Booth C (ed) Methods in microbiology, vol 4. Academic Press, London, pp 295–334
Xu Y, He J, Tian XP, Li J, Yang LL, Xie Q, Tang SK, Chen YG, Zhang S, Li WJ (2012) Streptomyces glycovorans sp. nov., Streptomyces xishensis sp. nov. and Streptomyces abyssalis sp. nov., isolated from marine sediments. Int J Syst Evol Microbiol 62:2371–2377
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA and whole genome assemblies. Int J Syst Evol Microbiol 67:1613–1617
Zhang J, Wang JD, Liu CX, Yuan JH, Wang XJ, Xiang WS (2014) A new prenylated indole derivative from endophytic actinobacteria Streptomyces sp. neau-D50. Nat Prod Res 7:431–437
Acknowledgements
This work was supported by a grant from King Mongkut’s Institute of Technology Ladkrabang Research Fund (KReF; KREF046301) to C.T. We thank the Actinobacterial Research Unit (ARU), Faculty of Science, King Mongkut’s Institute of Technology Ladkrabang, for laboratory supports.
Funding
The authors gratefully acknowledge the financial support from King Mongkut’s Institute of Technology Ladkrabang Research Fund (KReF; KREF046301) to C. T. and the Faculty of Science, King Mongkut’s Institute of Technology Ladkrabang to A. N. (RA/TA-2562-M-021).
Author information
Authors and Affiliations
Contributions
AN and CT carried out the experiments, analysed the data, and drafted manuscript. AN and CT contributed equally to this work. CS carried out fatty acid analysis. CT supervised the project. CI, PP, ST and ST corrected and reviewed the draft.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Ethics approval
This article does not contain any studies with human participants and/or animals performed by any of the authors. Formal consent is not required in this study.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Nammali, A., Intaraudom, C., Pittayakhajonwut, P. et al. Streptomyces endocoffeicus sp. nov., an endophytic actinomycete isolated from Coffea arabica (L.). Antonie van Leeuwenhoek 114, 1889–1898 (2021). https://doi.org/10.1007/s10482-021-01648-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10482-021-01648-x