Abstract
Infectious bronchitis (IB) is a highly contagious respiratory disease caused by a gammacoronavirus that has been circulating for many years in chickens in Bangladesh, resulting in significant economic losses. The aim of this study was to detect and characterize infectious bronchitis virus (IBV) from clinical outbreaks and surveillance samples. Real-time RT-PCR was used to detect IBV in pooled lung and tracheal tissue samples (n = 78), oropharyngeal swabs (n = 19), and pooled fecal samples (n = 13) from live-bird markets. Both respiratory and nephropathogenic forms of IB were suspected at necropsy (n = 7) from clinical outbreaks. Sequencing of hypervariable regions (HVR1-2 and HVR3) of the region of the spike gene (S) encoding the S1 subunit of five isolates revealed circulation of the Mass-like, QX-like, and 4/91-like genotypes of IBV in Bangladesh. Each genotype was extremely variable, as shown by separate clustering of the viruses in a phylogenetic tree and high nucleotide (nt) sequence divergence (38.8–41.2% and 25.7–37.4% in the HVR1-2 and HVR3 sequence, respectively). The unique mutation G65E was observed in each Mass-like isolate, and Y328S was observed in each 4/91-like Bangladeshi isolate. Three neutralizing epitope sites were predicted within the HVRs that differed significantly among the three genotypes. In addition, one Bangladeshi isolate carried fixed mutations at 294F and 306Y, like other pathogenic QX-like IBVs, which could affect epitopes involved in neutralization, facilitating virus circulation among vaccinated flocks. Therefore, continuous screening and genotype characterization will be necessary to track the epidemiology of IBV and control IB infection in Bangladesh.
Similar content being viewed by others
Introduction
Coronaviruses (CoVs) are continuously circulating in mammals and birds and pose a threat to livestock, companion animals, and humans. Avian coronaviruses (AvCoVs), including infectious bronchitis virus (IBV), belong to the genus Gammacoronavirus in the family Coronaviridae, order Nidovirales [21]. Infectious bronchitis (IB) is an acute, highly contagious disease caused by IBV that mainly infects the respiratory system but also affects other organs such as kidneys and the reproductive system of layer and breeder chickens of all ages worldwide [11]. AvCoVs also infect other domestic birds, such as turkeys, guinea fowl, quail, ducks, and migratory birds [7, 8, 13, 28]. IB in chickens is characterized predominantly by tracheal rales, coughing, sneezing, and thickened air sacs. The morbidity of IB is about 100%, but mortality may vary between 0% and 80%, depending on the age and immune status of the birds, strain of the virus, and involvement of concurrent pathogens [3, 32]. In layer birds, IB results in decreased egg production and reduced egg quality, and nephropathogenic IBV variants induce acute nephritis with polyuria evident by watery droppings [11]. Congested and swollen kidneys with deposition of urates are the important pathological findings in poultry infected with the nephropathogenic strain of IBV [1].
IBV has a single-stranded, non-segmented, linear positive-sense RNA genome of 27.6 kb. The genome has a 5′ capped end and a 3′ poly(A) tail and contains short untranslated regions (UTRs). Two overlapping large open reading frames (ORFs) encoding the replicase polyproteins 1a and 1b, which are the precursors of the viral RNA-dependent RNA polymerase (RdRp), occupy about two-thirds of the genome and are involved in RNA synthesis [17]. IBV encodes four structural proteins – spike (S), membrane (M), envelope (E) and nucleocapsid (N) – and 15 non-structural proteins (nsps) in addition to NSP1 [24], which is encoded within the large polyprotein 1ab. The largest structural protein, the S glycoprotein (an integral membrane protein), is cleaved by proteolysis at the highly basic furin consensus motif RRFRR/HRRR into S1 and S2 subunits, which facilitates attachment and entry of the virus into the host cell [39]. The S1 subunit contains three hypervariable regions: HVR1 (amino acid [aa] position 38-67), HVR2 (aa 91-141), and HVR3 (aa 274-387) [10, 22, 39], which are involved not only in viral infectivity but also contain at least five neutralization sites [20] and serotype-specific epitopes [9]. HVRs are highly variable, leading to numerous antigenic variants circulating globally [33]. Emergence of new variants of IBV occurs due to insertions, deletions, and substitutions in the genome and RNA recombination within the S1 gene. The S2 subunit of the S protein is responsible for fusion between viral and host cell membrane and contains a narrow stalk ectodomain, a short transmembrane segment, and a C-terminal domain [39].
Phylogenetic analysis based on IBV S1 gene sequences has revealed the existence of seven genotypes (GI-GVII) consisting of 35 genetic lineages, as well as inter-lineage recombinants [24]. Genotype GI includes 29 genetic lineages, whereas each of the other genotypes has only one lineage. The GI-1 and GI-12 lineages representing the old Massachusetts (Mass) and 793B lineages, respectively, have been found in all IB-endemic countries, while other lineages are found only in specific parts of the world. The 4/91-like and QX-like IBV strains belong to lineage GI-13 and GI-19, respectively, whereas the Israeli variant II and Egyptian variants I and II belong to the GI-23 lineage [14, 37]. IBV variants of the GI-23 lineage circulate in the Middle East and continue to spread in many countries in Africa, Asia, and Europe [24].
In Bangladesh, the poultry industry is an important source of employment and cheap animal proteins. However, this rapidly growing sector is continuously facing many challenges. The emergence and re-emergence of infectious viruses with higher virulence is occurring due to the constant natural evolution of various strains through recombination or mutation. Climatic changes and the frequent uses of live vaccines may be important reasons for this phenomenon. IB in Bangladesh has been reported in commercial poultry (layer, broiler and breeder farms), Sonali (cross-breed), and backyard chickens in association with other infectious diseases including Newcastle disease (ND), avian influenza (AI), infectious laryngotracheitis, chronic respiratory disease caused by mycoplasma, and egg drop syndrome 76 [5]. Several serosurveillance studies have confirmed the presence of IB in Bangladesh [4, 6, 15]. IBV strains within a geographical area are usually unique and distinct, although many countries share some common antigenic types of the virus [24]. Therefore, it is very important to determine the prevalence of IBVs in a region or country and to investigate the genetic characteristics of circulating strains in order to develop effective vaccines. In Bangladesh, live attenuated vaccines are being used on commercial poultry farms against IB infection without knowing the genetic characteristic of the circulating viruses. However, outbreaks of IB are reported frequently in Bangladesh due to poor vaccination strategies and vaccine mismatches [5]. A recent phylogenetic study revealed the presence of QX-like and 4/91-like strains of IBV in some areas of Bangladesh during 2014 to 2016 [5]. However, genetic and virological data for recent IBV strains in Bangladesh are lacking. Here, we describe the molecular detection and characterization of IBV from recent field outbreaks as well as samples from live-bird markets in Bangladesh.
Materials and methods
Sample collection
A total of 110 samples, including 78 tissue samples (lungs and trachea pooled for each) from natural field outbreaks, 19 oropharyngeal swabs, and 13 fecal samples (pooled from the surfaces or cages where each type of bird is sold) from live-bird markets (LBMs) from a surveillance study of respiratory pathogens, were obtained in the context of routine veterinary measures. Clinical outbreaks were recorded in 78 small-to-medium-size commercial and backyard layer and meat-type chicken flocks (broiler, Sonali, cockerel, and color bird) suffering from respiratory infections located in Mymensingh Division during 2020. The surveillance samples were from LBMs and apparently healthy commercial free-range duck flocks. Duck flocks were used to identify a gammacoronavirus other than IBV, because waterfowl act as carriers of different avian coronaviruses. Supplementary Table S1 contains detailed sample information. Tissues were homogenized to prepare a 20% w/v suspension in minimum essential medium (MEM) (Gibco by Life Technologies, Fisher Scientific, UK) containing a mixture of streptomycin and penicillin (50 µg/mL), and supernatant was collected [27], whereas swabs and faeces were collected directly in MEM without adding any serum supplement. The samples were kept at -20 °C until used.
Viral RNA extraction and detection by RT-qPCR
Viral RNA was extracted using a Monarch Total RNA Miniprep Kit (New England Biolabs Inc., USA) in accordance with manufacturer’s instructions. Single-step RT-qPCR assays were performed with previously designed unique oligonucleotide primers for the detection of the untranslated regions (5′-UTR) and a fragment of the polymerase gene of avian gammacoronaviruses (AvCoV) in a single assay [13]. The assay was carried out using a Luna Universal One-Step RT-qPCR Kit (New England Biolabs Inc., USA) containing SYBR Green reagents following a slightly modified previous protocol [28]. The final reaction volume was 12.5 μL, including 2.5 μL of RNA template, 5 μL of 2x RT-PCR SYBR reaction mix, 0.5 μL of Luna WarmStart RT Enzyme Mix, 2.5 μL of nuclease-free water, and 2 μL of IBV primer mix (10 pmol each). The RT-qPCR thermocycling conditions were 55 °C for 10 min (reverse transcription) and 95 °C for 2 min (initial denaturation), followed by 45 cycles at 95 °C for 10 s (denaturation) and 60 °C for 30 s (annealing and elongation) with reading of fluorescence in this step. Immediately after PCR, a melting curve analysis was performed with a continuous temperature increment of 0.5 °C/s between 65 °C and 95 °C. Samples yielding Cq values ≤ 32 with an appropriate melting temperature were considered positive for AvCoV RNA. Positive samples were further screened for IBV using probe-based (TaqMan) single-step RT-qPCR assays, using a Luna Universal Probe One-Step RT-qPCR Kit (New England Biolabs Inc., USA) and IBV-specific oligonucleotides [8]. All samples were screened for other respiratory viral pathogens, including avian influenza virus (AIV) and Newcastle disease virus (NDV) following a previously described protocol [28].
Virus isolation and identification
IBV RT-qPCR-positive samples were selected and prepared for inoculation of 10-day-old specific-pathogen-free embryonated chicken eggs by the allantoic route, using a standard protocol [27]. Inoculated eggs were incubated at 37 °C for 96 hours and candled daily to check the viability of the embryo. Allantoic fluid (AF) from dead and surviving embryos was harvested and retested to confirm the presence IBV RNA using an RT-qPCR assay. Samples with quantification cycle (Cq) values ≤ 26 and distinct amplification products in gel electrophoresis were subjected to amplification of the target gene by conventional RT-PCR.
Amplification of the spike gene region by conventional RT-PCR
HVR 1-2 and HVR 3 of the S1 gene of IBV were amplified by standard RT-PCR using specific oligonucleotide primers [2, 26]. A OneTaq One-Step RT-PCR Kit (New England Biolabs Inc., USA) was used to prepare a 25-μL reaction mixture containing: 3 μL of RNA template, 12.5 μL of 2X reaction buffer, 1 μL of each of the forward and reverse primers, 0.5 μL of OneTaq enzyme mix, and 7 μL of nuclease-free water. The thermocycling conditions for RT-PCR were 48 °C for 30 min (reverse transcription) and 94 °C for 1 min (initial denaturation), followed by 39 cycles at 94 °C for 15 s (denaturation), 30 s at 55 °C (annealing), and 40 s at 68 °C (extension), and then a final extension at 68 °C for 5 min.
Sequencing and phylogenetic analysis
Amplified RT-PCR products were purified using a FavorPrep GEL/PCR Purification Mini Kit (Favorgen, Taiwan) according to manufacturer’s instructions, and the DNA concentration was measured using a NanoDrop spectrophotometer (Thermo Scientific, USA). Gene sequencing was performed by a commercial service (Macrogen, South Korea) using the PCR primers in both directions. A BLAST (http://www.ncbi.nlm.nih.gov/BLAST) search was conducted for each raw sequence obtained. Sequences were edited and assembled using BioEdit software. Reference sequences of global IBV were downloaded from the NCBI database based on the selection criteria of homology, geographic location, and genotypic variations (Supplementary Table S2). Sequence comparisons were made and a maximum-likelihood (ML) phylogenetic tree was constructed using the Tamura-Nei model in the MEGA X program [23], using the ClustalW alignment algorithm [35] with 1,000 bootstrap replicates. Initial tree(s) for the heuristic search were obtained automatically by applying the neighbor-joining and BioNJ algorithms to a matrix of pairwise distances estimated using the maximum-composite-likelihood (MCL) approach and then selecting the topology based on log likelihood values. The sequences generated in this study were deposited in the GenBank database under the accession numbers listed in Supplementary Table S1.
Molecular analysis and structural epitope prediction of the S1 protein
After ClustalW alignment, the HVR1-2 and HVR3 regions of the S1 gene were converted to aa sequences, and the aa sequences of five Bangladeshi strains were compared with those of reference strains of different IBV genotypes. Furthermore, a 539-aa portion of the S1 protein (AFM46264.1 S1 protein) of a QX-like IBV strain was used to generate a model structure. The structure was modeled using the Swiss-Model repository's best-fit template (6cv0.1.A) of a cryoelectron microscopy structure of the IBV spike protein. Structural predictions of epitopes were made using the ElliPro antibody epitope prediction tool (http://tools.iedb.org/ellipro/) from the PDB file of the S1 model generated using Swiss-Model. Epitope structure predictions were performed using default parameters (a minimum score value of 0.5 and maximum distance of 6Å) [29]. Selected 3D epitope structures with significant scoring were viewed using the Jmol applet (http://www.jmol.org/).
Results
Clinicopathological characteristics
Commercial chicken flocks suffering from respiratory infections were chosen in the current study for clinicopathological examination. Clinical manifestations, mortality rates, and gross pathological observations varied among flocks and bird types, and also with involvement of coinfection. The mortality rate on the IBV-positive farms (n = 7) varied from 1% to 24%. The highest mortality rate (24%) was found in Sonali flocks, which were found to have mixed infections with NDV (RT-qPCR positive), and 16% in cockerel flocks, in which the mortality rate could have been elevated due to infection with mycoplasmas or other bacterial contamination, as suggested by gross pathology. Only 1-3% mortality was observed when IBV was the only infecting pathogen detected (Table 1). In general, the common clinical signs of the chickens with IB included respiratory distress in the form of tracheal rales, nasal discharge, or gasping. Table 1 summarizes the findings of a thorough investigation of seven IBV-positive cases from field outbreaks and five positive samples from a surveillance study of apparently healthy birds at LBMs (n = 2) and small-scale free-range duck farms (n = 3). The affected birds usually recovered within 10-15 days after applying symptomatic treatment and isolation of the affected birds from the healthy ones.
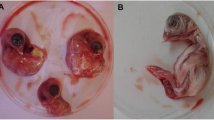
IBV-positive layer chickens consistently exhibited numerous lesions in the lungs, ova, liver, and kidney, while lesions in meat-type birds (broiler, Sonali, and cockerel) were restricted to the lungs, with the exception of hemorrhages in the kidneys in one Sonali chicken flock. At necropsy, lesions such as fragile lungs with congestion and hemorrhages, thickened and congested trachea, congested and fragile liver, hemorrhagic ova, and severe nephritis, were identified (Fig. 1). The lesions observed in the target organ were used to classify the pathogenic form in positive IBV cases found in this study (Table 1).
Identification of infectious bronchitis virus (IBV)
The results of RT-qPCR revealed that 12 samples were positive for avian gammacoronavirus, seven of which were from clinical outbreaks (n = 78), three from commercial free-ranging duck flocks (n = 19), and two from LBMs (n = 13) with positivity rates of 16%, 15%, and 15%, respectively. The three duck samples were positive in the AvCoV-specific RT-qPCR assay but negative in the IBV-specific probe-based RT-qPCR assay. Therefore, nine samples out of 110 were identified as IBV positive, resulting an overall prevalence of 8%. Mixed infection with AIV was observed in a single layer flock (LT 14), whereas a Sonali flock showed mixed infection with NDV (Table 1).
Characterization of IBV strains
Fragments of the S gene of IBV corresponding to HVR fragments (HVR1-2 and HVR3) of the S1 subunit were amplified from five IBV-positive samples (LT1, LT46, LT57, LBM1, and LBM5). The sequences obtained were aligned with sequences from selected global IBV strains, and those from the clinical outbreaks showed similarity to Mass-like (LT1 and LT46) and QX-like (LT57) IBV strains, whereas two sequences obtained from surveillance samples showed similarity to 4/91-like strains of IBV. The LT1 and LT46 isolates showed the highest nucleotide sequence similarity (HVR 1-2, 99.27% identity; HVR 3, 100% identity) to Mass-like strains from field isolates from China, Pakistan, Brazil, Poland, and Iran. The remaining field isolate, LT57, showed close similarity (HVR 1-2, 98.79-99.76% identity; HVR 3, 100% identity) to Chinese QX-like strains. On the other hand, the two IBV isolates from LMB (LMB1 and LBM5) showed 99.76-100% (HVR1-2) and 99.10-99.70% (HVR 3) nucleotide sequence identity to 4/91-like field strains from China.
Phylogenetic analysis demonstrated clustering of the five IBV sequences from Bangladesh. Both HVR1-2 and HVR3 phylogenies confirmed the existence of three genotypes – Mass-like, QX-like, and 4/91-like – in Bangladesh (Fig. 2). LT1 and LT46 clustered with Mass-like strains. The remaining field isolate, LT57, belonged to the QX-like genotype. On the other hand, the IBV isolates from LBMs (LBM1 and LBM5) grouped with 4/91-like strains.
Phylogenetic tree based on HVR1-2 (a) and HVR3 (b) sequences of the S1 gene. Isolates from the current study are indicated by a red triangle. Green circles (b) indicate sequences from Bangladesh from a previous study. Black circles indicate sequences from vaccine strains. All studied strains clustered with their respective genotypes, as shown in each tree in brackets. The evolutionary history was inferred using the maximum-likelihood method and the Tamura-Nei model [34] with 1000 bootstrap replicates. The percentage of trees in which the associated taxa clustered together is shown next to the branches. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. This analysis included 77 nucleotide sequences for HVR1-2 and 79 nucleotide sequences for HVR3. The final dataset included a total of 471 positions for HVR1-2 and 393 positions for HVR3. Evolutionary analyses were conducted in MEGA X [23].
The evolutionary distances among Bangladeshi strains were calculated in comparison to three well-established IBV genotypes (Mass-like, QX-like, and 4/91-like). Each genotypic group of Bangladeshi strains differed significantly from the others, as confirmed by pairwise divergence analysis of the HVR1-2 and HVR3 regions (Supplementary Table S3). Analysis of the HVR1-2 gene sequences revealed nucleotide distances of 0-41.2% among the five IBV isolates from Bangladesh, indicating a very high genetic divergence (Supplementary Table S3a). A similar trend, but with different divergence values, was observed with HVR3 (Supplementary Table S3b). The Bangladeshi IBV isolates showed high nucleotide sequence divergence (0-37.4%) among themselves, but they were closely related to the phylogenetically similar viruses.
Analysis of amino acid and epitope variations
Variations in amino acid (aa) sequences between different genotypes are quite common. We compared the aa sequences of the HVR1-2 and HVR3 regions of the five characterized strains from Bangladesh (Supplementary Table S4). Although the Bangladeshi field strains are closely related to members of their respective genotypes (Mass-like, QX-like, and 4/91-like), a few aa substitutions were observed. The two Mass-like strains (LT1 and LT46) from Bangladesh had glutamic acid (E) instead of glycine (G) at position 65 in the HVR1-2 region, but the HVR3 region was conserved. The HVR1-2 and HVR3 region of the QX-like strains had the largest number of aa variations, at two and eight different locations, respectively, while the Bangladeshi strain LT57 resembled the majority of the QX-like Chinese strains (Supplementary Table S4). Similar to the QX-like viruses, the HVR1-2 region of 4/91-like strains showed sporadic mutations at two sites, whereas in HVR3, two Bangladeshi 4/91-like strains (LBM1 and LBM5) showed a unique Y328S substitution.
Considering the high degree of genetic variation in the S1 subunit among different genotypes, we examined individual epitopes. A total of nine linear epitopes with a protrusion index (PI) above 0.7 (Supplementary Table S5) were found in the S1 subunit protein model. Interestingly, three of the nine epitopes were in the HVR1-2 and HVR3 regions. We therefore selected these three linear epitopes for Jmol structural visualization (Fig. 3a-c). Significant aa variations were observed in these three predicted epitopes in the three genotypes found in the current study, as shown in Figure 3. These variations presumably have the potential to affect the neutralizing activity of the virus. Two amino acid variations, 294F and 306Y, were found in the predicted linear epitope of HVR3 (Fig. 3c) in the two QX-like Bangladeshi IBV strains (LBM1 and LBM5).
Structural view of three predicted linear epitopes within the HVR1-2 (a and b) and HVR3 (c) regions of the S1 subunit of IBV. The epitope peptides are shown with the atoms of the backbone of the S1 subunit in yellow. The structure is based on a QX-like strain. The amino acid variations in the epitope sites of the Mass-like and 4/91-like Bangladeshi strains compared to the QX-like strain are shown in red. Protrusion index (PI) values are shown.
Discussion
This study was initiated to investigate the molecular epidemiology of IBV and characterize the currently circulating strains in commercial layer and meat-type chickens and domestic free-range ducks in Bangladesh. The current expansion of the poultry industry in Bangladesh, together with the national complex trading chain, international trade and movement, and introduction of vaccines to control the disease has coincided with the emergence and spread of new viral pathogens and strains [28, 31]. Considering the genetic and antigenic variability of IBV and the inadequacy of the vaccination program, characterization of circulating strains based on genotype and serotype is critical for preventing IBV infection. Cross-neutralization assays are commonly used to assess the serotype of IBV isolates. However, these assays are limited by the lack of standard positive sera. As a result, genotype classification based on the S1 gene has replaced serotype classification as the primary method of classifying IBV strains. Therefore, in the current study, we used two hypervariable regions (HVR1-2 and HVR3) of the S1 gene of IBV to identify the genotypes found in the field samples.
In Bangladesh, IBV is very common in major poultry-raising areas. The prevalence of IBV in the country varies among bird types, geographic locations, and seasons [4,5,6, 15]. In our study, we also detected IBV in a number of bird types, including commercial and backyard layers, broilers, Sonali, and cockerels. Surveillance of respiratory pathogens in duck populations also confirmed the presence of gammacoronaviruses other than IBV in the current study. Due to the small amount of viral RNA present, those samples were not used for further characterization. The mortality varied in clinical outbreaks in commercial layer and meat-type bird flocks and was increased by the presence of coinfections with avian influenza virus (AIV) and Newcastle disease virus (NDV). This study did not consider further avian viral or bacterial pathogens that are likely to complicate the overall clinical picture. The prevalence of IBV in different chicken breeds and considerable frequency of mixed infection with AIV and NDV have been reported in many countries in Asia and Africa [18, 19, 30, 36]. Here, in the clinical outbreaks, the respiratory form of the disease was relatively common and characterized by congested lungs, but we also detected the nephropathogenic form, indicated by the presence of severe nephritis (Fig. 1), in a layer (LT1) flock and a Sonali (LT46) flock, and viruses from two nephropathogenic lesions were characterized as Mass-like isolates. Another clinical case, LT57, was also confirmed as the respiratory form of the disease, and the virus was classified as QX-like. The two other positive cases (LBM1 and LBM5) identified using fecal samples from apparently healthy birds sold at LBMs were not included in the pathotype classification but were classified as the 4/91-like genotype. In phylogenetic analysis, the characterized strains clustered with their respective genotypes. Multiple pathotypes and genotypes have also been reported in China after an extensive long-term surveillance study [16].
The sampling year 2020 was very difficult due to the ongoing COVID-19 pandemic. Within the limited time frame for sampling, we identified two pathotypes and three genotypes of IBV strains. The QX-like and 4/91-like genotypes were reported previously in Bangladesh [5], but this is the first report to confirm the presence of the nephropathogenic Mass-like genotype in the country. All three identified genotypes are distinct from each other, as confirmed by phylogenetic analysis, divergence analysis, and amino acid (aa) sequence analysis. Our study also identified two unique substitutions: G65E in the HVR1-2 region of Mass-like strains and Y328S in the HVR3 region of 4/91-like strains. However, it is not clear when and where these mutations were introduced or their impact on virus pathogenicity. Therefore, it is necessary to continue surveillance and monitor the evolution of the virus.
The presence of vaccine strains in field outbreaks is an important consideration for sequence analysis, which can be challenging. According to the HVR1-2 and HVR3 phylogeny, it was observed that the Bangladeshi strains are closely related to field strains from China. Nobilis® MA5 (Mass-type) and Nobilis® IB 4-91 (4/91-type) live attenuated vaccines, manufactured by MSD Animal Health, have been used for decades in a fraction of the layer and breeder farms of Bangladesh. However, a large number of small-scale poultry farms remain unvaccinated. Due to poor vaccination coverage, IB continues to circulate in Bangladesh. Furthermore, the inclusion of various genotypes and the use of a live attenuated vaccine strains has increased the risk of recombination between the vaccine strains and pathogenic field strains (such as the QX-strain), which could result in a virulent recombinant strain, as reported in China and Egypt [16, 25]. Thus, recombination continues to be an important source of emergence of new variant strains of IBV. Genetic variations may result in changes in the tissue tropism and pathogenicity of the circulating viruses [12].
The presence of different genotypes and serotypes makes IBV prevention and control challenging. Although serotype classification was not performed in this study, in silico protein structural predictions were made that suggested that a number of the aa substitutions that were observed are in antigenic epitopes involved in virus neutralization that are characterized by their surface accessibility and antigenic reactivity with immunoglobulins [38] (Fig. 3). The presence of extremely variable aa residues in three predicted linear epitopes of each genotype may significantly reduce cross-protection and may lead to the generation of IBV escape mutants [40]. Therefore, in silico epitope prediction along with genome characterization may be useful for understanding the pathogenic potential of the circulating strains in a region.
Conclusions
Three IBV genotypes – Mass-like, QX-like, and 4/91-like IBV – were found in the current study, with a Mass-like IBV strain being identified for the first time in the country. Circulation of multiple genotypes coupled with unique mutations in the HVR1-2 and HVR3 regions in the Mass-like and 4/91-like strains differentiated the field strains from the vaccine strains. These strains produced respiratory and nephropathogenic infections with mild to moderate severity and significant mortality rates in layer and meat-type birds in Bangladesh. Coinfection with avian influenza virus may have public-health implications. Furthermore, virus-neutralizing epitopes in Bangladeshi strains were found to be highly variable, which could lead to limited protection with genotype-specific vaccines. However, continuous molecular identification and characterization of the circulating viruses along with experimental vaccination challenges could provide important information for selecting a suitable vaccine strain. Therefore, further investigation is required, and, subsequently, strengthening of biosecurity and other control programs should be implemented.
Data availability
The sequences analyzed in this study were downloaded from the GISAID (https://www.gisaid.org) database. The sequence metadata and other related documents generated for bioinformatics are available as supplemental files.
References
Abdel-Moneim AS, Afifi MA, El-Kady MF (2012) Emergence of a novel genotype of avian infectious bronchitis virus in Egypt. Arch Virol 157:2453–2457
Adzhar A, Gough RE, Haydon D, Shaw K, Britton P, Cavanagh D (1997) Molecular analysis of the 793/B serotype of infectious bronchitis virus in Great Britain. Avian Pathol 26:625–640
Ambali AG, Jones RC (1990) Early pathogenesis in chicks of infection with an enterotropic strain of infectious bronchitis virus. Avian Dis 34:809–817
Barua H, Biswas PK, Anwar MN, Dey BC, Debnath NC (2006) Serosurvey and isolation of infectious bronchitis virus in chickens reared in commercial and semi-scavenging systems. Bangladesh J Microbiol 23:114–117
Bhuiyan ZA, Ali MZ, Moula MM, Giasuddin M, Khan ZUM (2019) Prevalence and molecular characterization of infectious bronchitis virus isolated from chicken in Bangladesh. Vet World 12:909–915
Biswas PK, Barua H, Uddin GM, Biswas D, Ahad A, Debnath NC (2009) Serosurvey of five viruses in chickens on smallholdings in Bangladesh. Prev Vet Med 88:67–71
Brown PA, Touzain F, Briand FX, Gouilh AM, Courtillon C, Allée C, Lemaitre E, De Boisséson C, Blanchard Y, Eterradossi N (2016) First complete genome sequence of European turkey coronavirus suggests complex recombination history related with US turkey and guinea fowl coronaviruses. J Gen Virol 97:110–120
Callison SA, Hilt DA, Boynton TO, Sample BF, Robison R, Swayne DE, Jackwood MW (2006) Development and evaluation of a real-time Taqman RT-PCR assay for the detection of infectious bronchitis virus from infected chickens. J Virol Methods 138:60–65
Cavanagh D, Davis PJ, Cook JK, Li D, Kant A, Koch G (1992) Location of the amino acid differences in the S1 spike glycoprotein subunit of closely related serotypes of infectious bronchitis virus. Avian Pathol 21:33–43
Cavanagh D, Mawditt K, Adzhar A, Gough RE, Picault JP, Naylor CJ, Haydon D, Shaw K, Britton P (1998) Does IBV change slowly despite the capacity of the spike protein to vary greatly? Adv Exp Med Biol 440:729–734
Cavanagh D (2007) Coronavirus avian infectious bronchitis virus. Vet Res 38:281–297
Cavanagh D, Gelb J (2008) Infectious bronchitis. In: Saif YM, Fadly AM, Glisson JR, McDougald LR, Nolan NK, Swayne DE (eds) Diseases of poultry, 12th edn. Iowa State Press, Ames, pp 117–135
Chamings A, Nelson TM, Vibin J, Wille M, Klaassen M, Alexandersen S (2018) Detection and characterisation of coronaviruses in migratory and non-migratory Australian wild birds. Sci Rep 8:5980
Chen L, Xiang B, Hong Y, Li Q, Du H, Lin Q, Liao M, Ren T, Xu C (2021) Phylogenetic analysis of infectious bronchitis virus circulating in southern China in 2016–2017 and evaluation of an attenuated strain as a vaccine candidate. Arch Virol 166:73–81
Das S, Khan M, Das M (2009) Sero-prevalence of infectious bronchitis in chicken in Bangladesh. Bangladesh J Vet Med 7:249–252
Feng K, Wang F, Xue Y, Zhou Q, Chen F, Bi Y, Xie Q (2017) Epidemiology and characterization of avian infectious bronchitis virus strains circulating in southern China during the period from 2013–2015. Sci Rep 7:6576
Gorbalenya AE, Enjuanes L, Ziebuhr J, Snijder EJ (2006) Nidovirales: evolving the largest RNA virus genome. Virus Res 117:17–37
Hassan KE, Shany SA, Ali A, Dahshan AH, El-Sawah AA, El-Kady MF (2016) Prevalence of avian respiratory viruses in broiler flocks in Egypt. Poult Sci 95:1271–1280
Hassan KE, El-Kady MF, El-Sawah AAA, Luttermann C, Parvin R, Shany S, Beer M (2021) Respiratory disease due to mixed viral infections in poultry flocks in Egypt between 2017 and 2018: upsurge of highly pathogenic avian influenza virus subtype H5N8 since 2018. Transbound Emerg Dis 68:21–36
Kant A, Koch G, van Roozelaar DJ, Kusters JG, Poelwijk FA, van der Zeijst BA (1992) Location of antigenic sites defined by neutralizing monoclonal antibodies on the S1 avian infectious bronchitis virus glycopolypeptide. J Gen Virol 73:591–596
King AMQ, Lefkowitz EJ (2018) Changes to taxonomy and the International Code of Virus Classification and Nomenclature ratified by the International Committee on Taxonomy of Viruses (2018). Arch Virol 163:2601–2631
Koch G, Hartog L, Kant A, van Roozelaar DJ (1990) Antigenic domains on the peplomer protein of avian infectious bronchitis virus: correlation with biological functions. J Gen Virol 71:1929–1935
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549
Miłek J, Blicharz-Domańska K (2018) Coronaviruses in Avian species—review with focus on epidemiology and diagnosis in wild birds. J Vet Res 62:249–255
Moharam I, Sultan H, Hassan K, Ibrahim M, Shany S, Shehata AA, Abo-ElKhair M, Pfaff F, Höper D, El Kady M, Beer M, Harder T, Hafez H, Grund C (2020) Emerging infectious bronchitis virus (IBV) in Egypt: evidence for an evolutionary advantage of a new S1 variant with a unique gene 3ab constellation. Infect Genet Evol 85:104433
Naguib MM, El-Kady MF, Lüschow D, Hassan KE, Arafa AS, El-Zanaty A, Hassan MK, Hafez HM, Grund C, Harder TC (2017) New real time and conventional RT-PCRs for updated molecular diagnosis of infectious bronchitis virus infection (IBV) in chickens in Egypt associated with frequent co-infections with avian influenza and Newcastle Disease viruses. J Virol Methods 245:19–27
OIE (2014) Manual of diagnostic tests and vaccines for terrestrial animals, chapter 2.3.4. Avian influenza. Accessed 29 April 2021. World Animal Health Organization, Paris
Parvin R, Kabiraj CK, Mumu TT, Chowdhury EH, Islam MR, Beer M, Harder T (2020) Active virological surveillance in backyard ducks in Bangladesh: detection of avian influenza and gammacoronaviruses. Avian Pathol 49:361–368
Ponomarenko J, Bui HH, Li W, Fusseder N, Bourne PE, Sette A, Peters B (2008) ElliPro: a new structure-based tool for the prediction of antibody epitopes. BMC Bioinform 9:514
Promkuntod N (2016) Dynamics of avian coronavirus circulation in commercial and non-commercial birds in Asia—a review. Vet Q 36:30–44
Rahman M, Chowdhury EH, Parvin R (2021) Small-scale poultry production in Bangladesh: challenges and impact of COVID19 on sustainability. Ger J Vet Res 1:19–27
Raj GD, Jones RC (1996) Immunopathogenesis of infection in SPF chicks and commercial broiler chickens of a variant infectious bronchitis virus of economic importance. Avian Pathol 25:481–501
Sjaak de Wit JJ, Cook JK, van der Heijden HM (2011) Infectious bronchitis virus variants: a review of the history, current situation and control measures. Avian Pathol 40:223–235
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526
Thompson JD, Higgins DG, Gibson TJ, Clustal W (1994) Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22(22):4673–4680
Umar S, Teillaud A, Aslam HB, Guerin JL, Ducatez MF (2019) Molecular epidemiology of respiratory viruses in commercial chicken flocks in Pakistan from 2014 through to 2016. BMC Vet Res 15:351
Valastro V, Holmes EC, Britton P, Fusaro A, Jackwood MW, Cattoli G, Monne I (2016) S1 gene-based phylogeny of infectious bronchitis virus: an attempt to harmonize virus classification. Infect Genet Evol 39:349–364
Vita R, Overton JA, Greenbaum JA, Ponomarenko J, Clark JD, Cantrell JR, Wheeler DK, Gabbard JL, Hix D, Sette A, Peters B (2015) The immune epitope database (IEDB) 3.0. Nucleic Acids Res 43:D405-412
Wickramasinghe IN, van Beurden SJ, Weerts EA, Verheije MH (2014) The avian coronavirus spike protein. Virus Res 194:37–48
Yu L, Jiang Y, Low S, Wang Z, Nam SJ, Liu W, Kwangac J (2001) Characterization of three infectious bronchitis virus isolates from China associated with proventriculus in vaccinated chickens. Avian Dis 45:416–424
Acknowledgments
The authors are thankful for the technical assistance of Md. Shafiqul Islam and Md. Ripon, Department of Pathology, Bangladesh Agricultural University, Mymensingh, Bangladesh.
Funding
This research was funded by the Ministry of Science and Technology (BS-104), Government of Bangladesh, and The World Academy of Science (TWAS), Grant number no. 20-284 RG/BIO/AS_G – FR3240314166
Author information
Authors and Affiliations
Contributions
Conceptualization, supervision, resources, and project administration: RP and EHC. Methodology, investigation, validation, software, formal analysis, and visualization: RP, JAB, CKK and MN. Writing – original draft preparation: RP. Writing, review and editing, RP, JAB, MN, CKK and EHC. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical statement
This study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Institutional Review Board (or Ethics Committee) of Bangladesh Agricultural University (BAURES//ESRC/VET/07).
Additional information
Handling Editor: Zhenhai Chen.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Parvin, R., Begum, J.A., Nooruzzaman, M. et al. Circulation of three genotypes and identification of unique mutations in neutralizing epitopes of infectious bronchitis virus in chickens in Bangladesh. Arch Virol 166, 3093–3103 (2021). https://doi.org/10.1007/s00705-021-05227-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00705-021-05227-3