Abstract
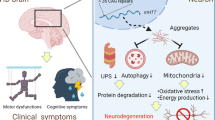
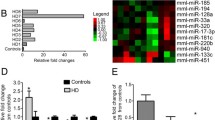
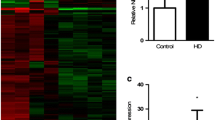
Huntington disease (HD) is the most common neurogenetic disorder caused by expansion of the CAG repeat in the HTT gene; nevertheless, the molecular bases of the disease are not fully understood. Non-coding RNAs have demonstrated to be involved in the physiopathology of HD. However, the role of circRNAs has not been investigated. The aim of this study was to identify the circRNAs with differential expression in a murine cell line model of HD and to identify the biological pathways regulated by the differentially expressed circRNAs. CircRNA expression was analyzed through a microarray, which specifically detects circular species of RNA. The expression patterns between a murine cell line expressing mutant Huntingtin and cells expressing wild-type Huntingtin were compared. We predicted the miRNAs with binding sites for the differentially expressed circRNAs and the corresponding target genes for those miRNAs. Using the target genes, we performed a function enrichment analysis. We identified 23 circRNAs differentially expressed, 19 downregulated and four upregulated. Most of the downregulated circRNAs derive from the Rere gene. The dopaminergic synapse, MAPK, and long-term depression pathways were significantly enriched. The three identified pathways have been previously associated with the physiopathology of HD. The understanding of the circRNA-miRNA-mRNA network involved in the molecular mechanisms driving HD can lead us to identify novel biomarkers and potential therapeutic targets. To the best of our knowledge, this is the first study analyzing circRNAs in a model of Huntington disease.


Similar content being viewed by others
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Rawlins MD, Wexler NS, Wexler AR, Tabrizi SJ, Douglas I, Evans SJ, Smeeth L (2016) The prevalence of Huntington’s disease. Neuroepidemiology. 46:144–153
Tabrizi SJ, Leavitt BR, Landwehrmeyer GB, Wild EJ, Saft C, Barker RA, Blair NF, Craufurd D, Priller J, Rickards H, Rosser A, Kordasiewicz HB, Czech C, Swayze EE, Norris DA, Baumann T, Gerlach I, Schobel SA, Paz E et al (2019) Targeting Huntingtin expression in patients with Huntington’s disease. N Engl J Med 380:2307–2316
Ross CA, Aylward EH, Wild EJ, Langbehn DR, Long JD, Warner JH, Scahill RI, Leavitt BR, Stout JC, Paulsen JS, Reilmann R, Unschuld PG, Wexler A, Margolis RL, Tabrizi SJ (2014) Huntington disease: natural history, biomarkers and prospects for therapeutics. Nat Rev Neurol 10:204–216
Saudou F, Humbert S (2016) The biology of huntingtin. Neuron. 89:910–926
Langbehn DR, Brinkman RR, Falush D, Paulsen JS, Hayden MR, International Huntington’s Disease Collaborative Group (2004) A new model for prediction of the age of onset and penetrance for Huntington’s disease based on CAG length. Clin Genet 65:267–277
Hatters DM (2008) Protein misfolding inside cells: the case of huntingtin and Huntington’s disease. IUBMB Life 60:724–728
Gusella JF, MacDonald ME (2009) Huntington’s disease: the case for genetic modifiers. Genome Med 1:80
Sharp PA (2009) The centrality of RNA. Cell. 136:577–580
Hsiao KY, Sun HS, Tsai SJ (2017) Circular RNA - new member of noncoding RNA with novel functions. Exp Biol Med (Maywood) 242:1136–1141
Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L (2014) Complementary sequence-mediated exon circularization. Cell. 159:134–147
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, Chen D, Gu J, He X, Huang S (2015) Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res 25:981–984
Maass PG, Glažar P, Memczak S, Dittmar G, Hollfinger I, Schreyer L, Sauer AV, Toka O, Aiuti A, Luft FC, Rajewsky N (2017) A map of human circular RNAs in clinically relevant tissues. J Mol Med (Berl) 95:1179–1189
Rybak-Wolf A, Stottmeister C, Glažar P, Jens M, Pino N, Giusti S, Hanan M, Behm M, Bartok O, Ashwal-Fluss R, Herzog M, Schreyer L, Papavasileiou P, Ivanov A, Öhman M, Refojo D, Kadener S, Rajewsky N (2015) Circular RNAs in the Mammalian brain are highly abundant, conserved, and dynamically expressed. Mol Cell 58:870–885
D'Ambra E, Capauto D, Morlando M (2019) Exploring the regulatory role of circular RNAs in neurodegenerative disorders. Int J Mol Sci 20:5477. https://doi.org/10.3390/ijms20215477
Holdt LM, Kohlmaier A, Teupser D (2018) Circular RNAs as therapeutic agents and targets. Front Physiol 9:1262
Scahill RI, Wild EJ, Tabrizi SJ (2012) Biomarkers for Huntington’s disease: an update. Expert Opin Med Diagn 6:371–375
Wyttenbach A, Swartz J, Kita H, Thykjaer T, Carmichael J, Bradley J, Brown R, Maxwell M, Schapira A, Orntoft TF, Kato K, Rubinsztein DC (2001) Polyglutamine expansions cause decreased CRE-mediated transcription and early gene expression changes prior to cell death in an inducible cell model of Huntington’s disease. Hum Mol Genet 10:1829–1845
Zhang B, Kirov S, Snoddy J (2005) WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res 33:W741–W748
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504
Yanagisawa H, Bundo M, Miyashita T, Okamura-Oho Y, Tadokoro K, Tokunaga K, Yamada M (2000) Protein binding of a DRPLA family through arginine-glutamic acid dipeptide repeats is enhanced by extended polyglutamine. Hum Mol Genet 9:1433–1442
Zoltewicz JS, Stewart NJ, Leung R, Peterson AS (2004) Atrophin 2 recruits histone deacetylase and is required for the function of multiple signaling centers during mouse embryogenesis. Development. 131:3–14
Fregeau B, Kim BJ, Hernández-García A, Jordan VK, Cho MT, Schnur RE, Monaghan KG, Juusola J, Rosenfeld JA, Bhoj E, Zackai EH, Sacharow S, Barañano K, Bosch DGM, de Vries BBA, Lindstrom K, Schroeder A, James P, Kulch P et al (2016) De novo mutations of RERE Cause a genetic syndrome with features that overlap those associated with proximal 1p36 deletions. Am J Hum Genet 98:963–970
Nagafuchi S, Yanagisawa H, Sato K, Shirayama T, Ohsaki E, Bundo M, Takeda T, Tadokoro K, Kondo I, Murayama N (1994) Dentatorubral and pallidoluysian atrophy expansion of an unstable CAG trinucleotide on chromosome 12p. Nat Genet 6:14–18
Koch ET, Raymond LA (2019) Dysfunctional striatal dopamine signaling in Huntington’s disease. J Neurosci Res 97:1636–1654
Gamez J, Lorenzo-Bosquet C, Cuberas-Borrós G, Carmona F, Hernández-Vara J, Castilló J, Castell-Conesa J (2010) Does reduced [(123)I]-FP-CIT binding in Huntington’s disease suggest pre-synaptic dopaminergic involvement? Clin Neurol Neurosurg 112:870–875
Paoletti P, Vila I, Rifé M, Lizcano JM, Alberch J, Ginés S (2008) Dopaminergic and glutamatergic signaling crosstalk in Huntington’s disease neurodegeneration: the role of p25/cyclin-dependent kinase 5. J Neurosci 28:10090–10101
Sørensen SS, Nygaard AB, Christensen T (2016) miRNA expression profiles in cerebrospinal fluid and blood of patients with Alzheimer’s disease and other types of dementia - an exploratory study. Transl Neurodegener 5:6-016-0053-5 eCollection 2016
Zhu J, Xu X, Liang Y, Zhu R (2021) Downregulation of microRNA-15b-5p Targeting the Akt3-Mediated GSK-3β/β-catenin signaling pathway inhibits cell apoptosis in Parkinson’s disease. Biomed Res Int 2021:8814862
Keshet Y, Seger R (2010) The MAP kinase signaling cascades: a system of hundreds of components regulates a diverse array of physiological functions. Methods Mol Biol 661:3–38
Seternes OM, Kidger AM, Keyse SM (2019) Dual-specificity MAP kinase phosphatases in health and disease. Biochim Biophys Acta, Mol Cell Res 1866:124–143
Taylor DM, Moser R, Régulier E, Breuillaud L, Dixon M, Beesen AA, Elliston L, Silva Santos Mde F, Kim J, Jones L, Goldstein DR, Ferrante RJ, Luthi-Carter R (2013) MAP kinase phosphatase 1 (MKP-1/DUSP1) is neuroprotective in Huntington’s disease via additive effects of JNK and p38 inhibition. J Neurosci 33:2313–2325
Apostol BL, Illes K, Pallos J, Bodai L, Wu J, Strand A, Schweitzer ES, Olson JM, Kazantsev A, Marsh JL, Thompson LM (2006) Mutant huntingtin alters MAPK signaling pathways in PC12 and striatal cells: ERK1/2 protects against mutant huntingtin-associated toxicity. Hum Mol Genet 15:273–285
Ha J, Kang E, Seo J, Cho S (2019) Phosphorylation dynamics of JNK Signaling: effects of dual-specificity phosphatases (DUSPs) on the JNK pathway. Int J Mol Sci 20:6157. https://doi.org/10.3390/ijms20246157
Ahmed T, Zulfiqar A, Arguelles S, Rasekhian M, Nabavi SF, Silva AS, Nabavi SM (2020) Map kinase signaling as therapeutic target for neurodegeneration. Pharmacol Res 160:105090
Nithianantharajah J, Hannan AJ (2013) Dysregulation of synaptic proteins, dendritic spine abnormalities and pathological plasticity of synapses as experience-dependent mediators of cognitive and psychiatric symptoms in Huntington’s disease. Neuroscience. 251:66–74
Ferrante RJ, Kowall NW, Richardson EP Jr (1991) Proliferative and degenerative changes in striatal spiny neurons in Huntington’s disease: a combined study using the section-Golgi method and calbindin D28k immunocytochemistry. J Neurosci 11:3877–3887
Raymond LA, André VM, Cepeda C, Gladding CM, Milnerwood AJ, Levine MS (2011) Pathophysiology of Huntington’s disease: time-dependent alterations in synaptic and receptor function. Neuroscience. 198:252–273
Acknowledgments
The authors acknowledge to the Consejo Nacional de Ciencia y Tecnologúa (CONACYT) and to David Rubinsztein from the Cambridge Institute of Medical Research for kindly donate the cell line.
Funding
This research was funded by Consejo Nacional de Ciencia y Tecnologúa (CONACYT) grant number FOSISS-273213
Author information
Authors and Affiliations
Contributions
Research project conception: Alejandra Camacho-Molina, Alberto Hidalgo-Bravo. Execution of experimental procedures: Ernesto Marfil-Marin, Mónica Santamaría-Olmedo, Adriana PerezGrovas-Saltijeral. Data analysis: Margarita Valdes-Flores, Adriana Ochoa-Morales, Aurelio Jara-Prado, Rosalba Sevilla-Montoya, Alejandra Camacho-Molina, Alberto Hidalgo-Bravo. Interpretation of results: Adriana Ochoa-Morales, Aurelio Jara-Prado, Santamaría-Olmedo, Adriana PerezGrovas-Saltijeral, Margarita Valdes-Flores. Draft of the manuscript: Ernesto Marfil-Marin, Rosalba Sevilla-Montoya, Alejandra Camacho-Molina, Alberto Hidalgo-Bravo. All authors reviewed and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Conflicts of interest
The authors declare no competing interests.
Ethics approval
This article employed a modified cell line derived from the organism Rattus norvegicus obtained from the University of Cambridge. No human material was used in this study. This work does not qualify for ethics approval according to the Institutional Ethics Committees from the participant institutions.
Cell lines
A cell line derived from the organism Rattus norvegicus (RRID:CVCL_0481) was obtained from David Rubinsztein from the Cambridge Institute of Medical Research.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
About this article
Cite this article
Marfil-Marin, E., Santamaría-Olmedo, M., PerezGrovas-Saltijeral, A. et al. circRNA Regulates Dopaminergic Synapse, MAPK, and Long-term Depression Pathways in Huntington Disease. Mol Neurobiol 58, 6222–6231 (2021). https://doi.org/10.1007/s12035-021-02536-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-021-02536-1