Abstract–
Acute lung injury (ALI) is a type of serious clinical syndrome leading to morbidity and mortality. However, the precise pathogenesis of ALI remains elusive. Here, we implemented an integrative meta-analysis of six GEO microarray studies with 76 samples in the ALI mouse model. A total of 958 differentially expressed genes (DEGs) were identified in LPS relative to normal samples. Then, a network-based meta-analysis was used to mine core DEGs and to unfold the interactions among these genes. We found that Ebi3 was the top upregulated genes in the LPS-induced ALI. GO, KEGG, and GSEA analyses were performed for functional annotation. qRT-PCR revealed augmented expression of six candidate genes (Stat1, Syk, Jak3, Rac2, Ripk1, and Traf6) in the established ALI mouse model with LPS exposure. Taken together, our study investigated comprehensively hub DEGs and their networks for LPS-stimulated ALI, which might afford an additional approach to determine biomarkers and therapeutic targets and explore the molecular pathophysiology toward ALI.
Similar content being viewed by others
INTRODUCTION
Acute lung injury (ALI) is one of the severe clinical complications distinguished by pulmonary vascular permeability [1]. Meanwhile, ALI exhibits predisposing and precipitating factors, such as acute pneumonia, acute pancreatitis, and severe trauma, and acute pancreatitis, and ultimately develops into acute respiratory distress syndrome (ARDS) [2]. To make matters worse, ALI and ARDS are linked to many destructive clinical disorders with high morbidity and mortality rates [3]. Lipopolysaccharide (LPS) is the main ingredients of the cell wall in gram-negative bacteria which acts as the pathogenic infection resulting in ALI [4]. Triggered by LPS stimulation, inflammatory cytokines, such as tumor necrosis factor-α (TNF-α) and interleukin-1β (IL-1β), might facilitate ALI development [5, 6] Tremendous core genes have been characterized to date and deepen our understanding of the pathogenesis of ALI [7,8,9]. Nevertheless, the immunological mechanism is still unclear.
Stunning advances in high-throughput technologies and bioinformatic mining have produced huge volumes of data. Conventional gene expression analysis generally focuses on a specific gene, discounting the effect of interactions among individual genes. Thereby, efficient and robust tools are the key to integrate available data and provide insight into complex diseases. Recently, a comprehensive approach is constructed using protein–protein interaction (PPI) data to interpret the interactive patterns across multiple datasets. This network meta-method has been developed as a feasible way to analyze large gene profiles and successfully applied in the health bioinformatics field [10,11,12].
To characterize hub genes and explore the molecular mechanisms of LPS-treated ALI, we built a compendium of genes related to ALI using gene expression profiles from six microarray studies. We then used the above PPI networked comprehensive analysis to detect the hub genes and functional modules. The ALI mice model was performed by intratracheally instilling with LPS. Seven candidate genes were confirmed using an established mouse model of ALI and qRT-PCR validation. The results pave the way for future research into the pathophysiology of ALI, which might influence far-reaching individual anti-ALI treatment. Despite a lot of putative biomarkers were yielded by screening a large amount of data, determining the functional effect of each gene will require in-depth verification in animal models and clinical samples.
MATERIAL AND METHODS
Microarray Data Processing
To mine the related genes of LPS-induced ALI in mice, gene expression microarray studies were derived from the GEO (Gene Expression Omnibus, http://www.ncbi.nlm.nih.gov/geo) repository by following strategies: (1) keyword search: “LPS-induced ALI” and “Mus musculus,” “lung,” and “expression profiling by array”; (2) it contained at least three specimens per group. Consequently, six available profiles (GSE102016, GSE2411, GSE16409, GSE104214, GSE17355, and GSE18341) met the inclusion criteria and were selected for the next analyses [13,14,15,16,17,18]. Detailed information (GEO ID, platform information, and sample accession) of these datasets is summarized in Table 1.
Integrated Multi-microarray Analysis
The integrated microarray analysis was conducted through a visual analytics platform NetworkAnalyst 3.0 [19]. Firstly, all gene probes were converted to Entrez ID. Secondly, we preprocessed the expression profiles by log2 transformation and auto-scaling before quantile normalization. For the meta-analysis based on NetworkAnalyst 3.0, three analytics methods were employed including the Fisher method, fixed-effect model (FEM), and vote-counting (combined p-value < 0.05 or vote counts ≥ 2 were considered to be significant) after adjusting study batch effect. Combined p-value < 0.05 or vote counts ≥ 2 were considered to be significant. In FEM, the estimated effect size (ES) in each group is assumed to come from an underlying true effect size plus measurement error. Finally, the common DEGs defined by above three methods were regarded as the final candidate genes.
Functional Enrichment Analyses of DEGs
According to the hypergeometric distribution algorithm, GO (Gene Ontology, http://www.geneontology.org/) enrichment analyses divided into biological process (BP), molecular function (MF), and cell component (CC) were implemented using “enrichGO” function of “clusterProfiler” R package [20]. To simplify enriched GO results, we then applied “enricher” function to reduce redundant GO terms (The threshold parameter was set by default to “cutoff = 0.7, by = “p.adjust”, select_fun = min”). Furthermore, the pathways of the identified proteins were classified via “enrichKEGG” function of “clusterProfiler” package for KEGG (Kyoto Encyclopedia of Genes and Genomes) annotation.
To test a set of related genes in coordinated way, GSEA (Gene Set Enrichment Analyses) of GO and KEGG were enforced by “gseGO” and “gseKEGG” functions of “clusterProfiler” R package, separately. The Hallmarks gene set was downloaded from MSigDB v7.1 (Molecular Signatures Database, https://www.gsea-msigdb.org/gsea/msigdb/index.jsp). P-value < 0.05 were considered as statistically significance.
Network-Based Meta-analysis and Extracting Co-expressed PPI Modules
A network-based analysis was performed using STRING search function in NetworkAnalyst website (parameters were set to “confidence score cutoff = 900” and “require experimental evidence”). The extended network construction was restricted to include only the original seed proteins. Thus, those with a high level of confidence were retained and regraded as valid interactions. Besides, PPI networks of core seven DEGs were predicted via the STRING v10.5 (Search Tool for the Retrieval of Interacting Genes/Proteins, http://www.string-db.org/). Proteins that linked to each other were detected based on corresponding gene neighborhood, co-occurrence, co-expression, protein homology, experimental determination, curated databases, and text mining.
The Murine Model Establishment of ALI and qRT-PCR Validation
The regents used and the detailed procedures of the murine model establishment and qRT-PCR were performed as before [8]. The H&E immunohistological staining images of the control group and the ALI group are shown in Figure S1. All experiment protocols conformed to the guidelines of the China Council on Animal Care and Use. These animal studies were approved by the Institutional Animal Research Committee of Union Hospital. Mice were randomly allocated into ten control samples and ten LPS samples and then feeding a week of mice to adapt the environment. Control mice were exposed to an aerosol of phosphate buffer saline (PBS) alone. For acute LPS exposure, mice were exposed to PBS containing 0.5 mg/mL LPS for 2 h, in a custom-built cuboidal chamber. The LPS solution was aerosolized with a constant output ultrasonic nebulizer (model: 402B, Yuwell, China) at a flow rate of 30 ml/h. LPS was purchased from Sigma-Aldrich (extracted from Escherichia coli O55: B5, L2880). The chamber was 18 cm long, 12 cm wide, and 12 cm high. The 2−ΔΔCt method was used, and the PCR reactions were normalized to the GAPDH gene. All primers were sourced from PrimerBank (https://pga.mgh.harvard.edu/primerbank/). The reverse transcription step of qRT-PCR was performed by the UEIris RT mix with DNase (All-in-One) (US Everbright® Inc., Suzhou, China). The qRT-PCR reaction (10 µL) was formulated by the 2 × SYBR Green qPCR Master Mix (US Everbright® Inc., Suzhou, China). All qRT-PCRs were carried out on a CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA). The primer sequences are provided in the Supplementary Materials (Table S1).
Statistical Analysis
The Benjamini–Hochberg application of the FDR (false discovery rate) was applied to correct the p-value. Significantly enriched GO terms and KEGG pathways were distinguished using hypergeometric tests. Statistical analysis of significant differences between groups in qRT-PCR was complied with one-way ANOVA by Prism 7 software.
RESULTS
A Compendium of the Eligible Microarrays in This Study
Under our inclusion criteria, a gene expression compendium was finally built using six independent studies (GSE18341, GSE104214, GSE17355, GSE102016, GSE16409, and GSE2411) from the GEO database. In GSE18341, juvenile (21 day) and adult (16 week) C57BL/6 mice were treated with inhaled LPS (20 mL of 0.1 mg/mL) for 30 min in a sealed aerosol chamber and then spontaneous breathing. Untreated age-matched controls acted as comparison groups. In GSE104214, the experimental group was treated with 5 μg of intranasal LPS at 8 and 24 h. Controls received intranasal PBS. As for GSE17355, injury peaked at day 4 and is almost completely resolved by day 10 in wild type C57BL/6 mice in LPS model of ALI. Total RNA was isolated from mouse lung at times 0, 1, 4, and 10 days following LPS treatment of wild type. In GSE102016, mice were intranasally treated with LPS (20 μg/mouse) to induce pulmonary inflammation and all lung samples were collected after 72 h. In GSE16409, samples were from control or LPS exposed (1.5, 6, and 12 h post exposure). Twenty-four C57/B6 male mice were randomized to control and LPS groups in GSE2411. A total of 47 LPS-challenged and 29 normal mice were included in this study (Table 1). Then, we extracted and annotated these six GEO data, yielding a compilation of 2030 genes from 76 samples totally. The entire work flow in this study is depicted in Figure S2.
Overlap of Differentially Expressed Genes Among Datasets
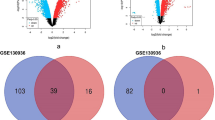
A comprehensive analysis across studies was performed by computing the DEGs per dataset and assessing the overlap of the significant results. As shown in Fig. 1A, 3400, 1392, 653, 194, 94, and 13 DEGs were generated in GSE18341, GSE104214, GSE17355, GSE102016, GSE16409, and GSE2411, respectively. Besides, seven overlapped genes (Ifi44, Tnip1, Oasl1, Casp4, Ccl12, Zbp1, and Cxcl13) were tightly associated with LPS among at least five datasets (Fig. 1A). Among them, caspase-4 (Casp4) acts as an innate immune receptor for cytoplasmic LPS [21]. And the inflammatory cytokines and chemokines Ccl12 and Cxcl13 have been reported to participate in the pathogenic mechanism of acute lung injury (ALI) [22]. These findings implied that these three genes in the five datasets were closely related with the LPS host response in mice.
Comprehensive analysis of six GEO compendium. (A) Significant ALI-associated genes in six independent studies of the ALI compendium. The upset plot showed the DEG count in each group. The black dots and lines between the dots represented DEGs in which GEO dataset. The bars represented number of DEGs in the group. (B) Venn diagram of DEGs identified from the meta-analysis using Fisher’s method, the vote counting method, and a fixed-effect model. (C) Heat map representation of the top 30 upregulated DEGs across different microarrays identified from the meta-analysis (row-wise comparison). The color of boxes varying from blue to red represented the combined effect size value of DEG in each sample.
Comprehensive Analysis of GEO Microarrays
Next, we performed meta-analyses through NetworkAnalyst 3.0 platform. The merged data of this analysis is tabulated in Table S2. After employing three meta-analysis methods, we validated 1703, 1316, and 996 DEGs by the Fisher method, FEM, and vote counting, respectively. Of those, 958 DEGs were identified via all three methods (Fig. 1B; Table S3). Thereinto, mRNA levels of 470 (49.1%) DEGs elevated and 488 (50.9%) DEGs were lower in LPS group in comparison with the control. Two heat map visualization of the top 30 up- and downregulated DEGs across the different studies is displayed in Fig. 1C and Figure S3, respectively. In the overlapping DEGS across the six microarray cohorts, Ebi3 was the top upregulated gene followed by F10, and Fmo3 was the most prominently downregulated gene.
Functional Enrichment and GSEA Analyses of DEGs
To exploit potential features of DEGs, we analyzed GO function and KEGG pathway enrichment. The top 5 most enriched KEGG pathways of DEGs were involved in osteoclast differentiation, fc gamma R-mediated phagocytosis, MAPK signaling pathway, fluid shear stress and atherosclerosis, and leishmaniasis (Fig. 2). Moreover, we were able to obtain a global perspective of the changes in gene expression patterns In GO enrichment analysis (Table S4). In MF term, DEGs were centered on “ubiquitin-like protein ligase binding,” “coenzyme binding,” and “GTPase activity.” As for the BP category, the core DEGs were significantly enriched related to “leukocyte proliferation,” “cytokine-mediated signaling pathway,” and “response to oxidative stress.” In addition, DEGs were enriched in the CC category focused on “membrane region,” “actin cytoskeleton,” and “NADPH oxidase complex” (Fig. 3).
In GSEA analysis, the gseGO outcomes showed that DEG expressions were mainly involved in response to a chemical, the immune system process, and response to an external stimulus (Fig. 4A). As for gseKEGG results, DEGs were associated with the TNF, IL-17, and C-type lectin receptor signaling pathway (Fig. 4B). Of these, the signaling pathways of cytokine-mediated, TNF, and IL − 17 were reported to accumulate in the lung epithelial cells within acute inflammation and serious viral infection [23]. Since ALI is characteristic of severe inflammation of lungs resulting from uncontrolled inflammatory immune response, the outcomes implied that these DEGs might play essential roles in the pathogenesis and progression of ALI/ARDS. Given the functional enrichment of these DEGs, we delved further into the results in an integrative meta-analysis.
Analysis of PPI Networks of DEGs
We searched for hub genes via NetworkAnalyst which might be useful as biomarkers and therapeutic targets for ALI. The “first-order” PPI network for ALI was difficult to interpret and navigate due to it yielded 3055 nodes and 7107 connection edges. To better present the PPI network, “zero order” network analysis was conducted possessing four subnetworks (at least 5 nodes), including one big subnetwork (subnetwork1 contained 223 nodes and 423 connection edges) and three smaller ones (subnetwork2–4; Table S5). We used Cytoscape software to better visualize the subnetwork1 (Fig. 5). DEGs within the subnetwork1 were ranked by the betweenness, and the top 7 DEGs (Stat1, Syk, Jak3, Rac2, Ripk1, Traf6, and Mapk3) were regarded as hub genes whose node degrees ≥ 10. Among them, Mapk3 was the most highly ranked key gene (degree = 12; betweenness = 3910.56) among the downregulated DEGs. Meanwhile, Stat1 was the centermost upregulated gene (degree = 23; betweenness = 10,125.24) followed by Syk (degree = 21; betweenness = 2883.29).
Module visualization and identification of hub genes. Visualization of the gene co-expression network of the DEGs was generated using Cytoscape software. The color of dot represented combined ES of DEGs, which varied from blue to red. Seven hub DEGs were highlighted by diamond dots while others were represented by circles.
Verification of Functional Roles Using an Established Mouse Model
To next confirm the candidate 7 core DEGs (Stat1, Syk, Jak3, Rac2, Ripk1, Traf6, and Mapk3) screened with this network-based approach, we employed an established ALI mice model through intratracheal instillation of LPS and utilized qRT-PCR validation. The results revealed that the relative mRNA levels for Stat1, Syk, Jak3, Rac2, Ripk1, and Traf6 were increased, while Mapk3 was lower expressed in the LPS group than the control (Fig. 6A–G; Table S6). Except those in Traf6 (p-value = 0.1544) and Mapk3 (p-value = 0.2156), LPS vs control comparisons in other genes showed the significant differences (p-value < 0.05). Overall, the qRT-qPCR results were in accord with our integrative meta-analysis, suggesting the critical role of the 7 key DGEs that might play in the pathogenic mechanism of ALI/ARDS. We subsequently tested whether these DEGs had an intrinsic relationship in ALI via analyzing the STRING database. We noticed that Traf6 had the most significant connections with other genes followed by Stat1 (Fig. 6H).
(A–G) Expression levels of seven core genes (Stat1, Syk, Jak3, Rac2, Ripk1, Traf6, and Mapk3) in the lung of established mouse model. GAPDH was used as an internal control gene. Experiments were performed at least in triplicates. ****, ***, **, *, and ns represent p-value < 0.0001, < 0.001, < 0.01, < 0.05, and no difference, respectively. (H) Functional links among core genes determined by STRING database.
DISCUSSION
ALI and ARDS are correlated with short-term and long-term syndromes, including physical and cognitive impairments [24, 25], as well as poor clinical conditions outcomes with mortality rates as high as 35–55% [26], whereas the underlying molecular mechanism of ALI/ARDS is far from being understood. In the present study, to reconnoiter the promising gene changes and key mediators that occur in ALI, we built a transcriptional compendium of the genes concerning LPS-induced ALI from six microarray studies that covered 76 tissues in total. Ultimately, 958 overlapping DEGs were confirmed and found to be enriched in several known terms of GO and KEGG pathways related to inflammatory responses and necroptosis. A PPI network-based analysis was used to evaluate the relationship between these genes. This analysis suggested these genes interrelated with four significant subnetworks. We detected 7 hub genes (Stat1, Syk, Jak3, Rac2, Ripk1, Mapk3, and Traf6) with at least 10 node degrees. Among them, Stat1 was the centermost over-expressed gene, followed by Syk. Mapk3 was the most clearly downregulated gene. We verified mRNA levels of these seven genes between LPS-challenged mice and normal mice. These outcomes demonstrated that Stat1, Syk, Jak3, Rac2, Ripk1, and Traf6 were markedly increased in an established aging model, as well as Mapk3 was decreased.
The innate immune response exerts a profound impact on the pathogenesis of ALI [27]. Miscellaneous signal transduction pathways partake in mediating lung inflammatory responses, such as the JAK/STAT, NF-κB, and MAPK signaling pathways [28]. Among them, the JAK/STAT pathway are critical determinants of initiating adaptive immune responses and constraining inflammatory responses eventually [29]. Janus tyrosine kinase 3 (JAK3), one of the JAK family members, is an attractive selective target for the treatment of immune-mediated disorders [30]. Inhibiting JAK3 reduces the hyperproduction of cytokine/chemokine [31]. STAT1 is an important component of inflammation and tumorigenesis. Recent studies have demonstrated that the downregulation of STAT1 might inhibit lung cell apoptosis in ALI [32]. As for spleen tyrosine kinase (SYK), it modulates inflammatory signaling in various immune cells via classical immunoreceptors [33]. LPS results in increasing Syk expression in neutrophils and gamma delta (γδ) T cells directed to inflammatory process of ALI [34].
Neutrophils, dendritic cell (DC)s, and macrophages are associated with multiple immunological processes and tissue injury by initiating inflammation during ALI [35]. As a member of the Rho GTPase subfamily, RAC2 acts as a necessary regulator of neutrophil degranulation. In alveolar macrophages and neutrophils, lung injury in response to immune complex deposition is reliant on Rac2 [36]. MAPK3 is a negative moderator of DC and is necessary for the induction of T cell anergy towards an inflammatory phenotype [37]. Tumor necrosis factor receptor–associated factor 6 (TRAF6) is a decisive adaptor regulating Toll-like receptors (TLRs), whose polyubiquitination leads to mediate pro-inflammatory cytokines production in ALI [38]. Moreover, necroptosis, a cell death form modulated by the RIPK1-RIPK3-MLKL signaling, has been implicated in the close relationship between necroptosis and ALI/ARDS [39, 40]. As cellular signaling molecules, receptor-interacting protein kinases (RIPKs) are critical for homeostatic signaling in many disease processes [41]. The activation of RIPK1/3-dependent necroptosis would result in ALI/ARDS and even death [42].
In summary, as mentioned above, Stat1, Syk, Jak3, Rac2, Ripk1, and Traf6 were the central upregulated genes in our network analysis. The expression of those DEGs was higher in LPS-induced pathological injury in the lung than control counterparts. The enhanced DEGs may be critical factors in attenuating LPS-stimulated ALI/ARDS. These observations coincided with the earlier reports proving our analyses are reliable and practical. On the other hand, some DEGs like Ebi3 and F10 have been described previously in many other illnesses [43,44,45], but their regulatory functions in ALI/ARDS have not been fully known. Additionally, this configuration of network analysis may serve as a novel tool to screen for corresponding potential signatures in an attempt to fill the gaps of knowledge about properties and pathogenesis of ALI. To address the limitations in this study, further research using knockout gene mice for each DEGs is indispensable and in urgent need. It will facilitate us to get better acquainted with its role of LPS in aggravating ALI/ARDS.
Availability of Data and Materials
All data generated and described in this article are available from the corresponding web servers, and are freely available to any scientist wishing to use them for noncommercial purposes, without breaching participant confidentiality.
Change history
13 November 2021
A Correction to this paper has been published: https://doi.org/10.1007/s10753-021-01584-y
Abbreviations
- ALI:
-
Acute lung injury
- ARDS:
-
Acute respiratory distress syndrome
- DEGs:
-
Differentially expressed genes
- LPS:
-
Lipopolysaccharide
- IL-1β:
-
Interleukin-1β
- TNF-α:
-
Tumor necrosis factor-α
- NF-κB:
-
Nuclear factor kappaB
- GEO:
-
Gene expression omnibus
- PPI:
-
Protein-protein interaction
- VSN:
-
Variance stabilizing normalization
- FDR:
-
False discovery rate
- FEM:
-
Fixed effect model
- ES:
-
Effect size
- MSigDB:
-
Molecular Signatures Database
- GSEA:
-
Gene Set Enrichment Analyses
- GO:
-
Gene ontology
- BP:
-
Biological process
- MF:
-
Molecular function
- CC:
-
Cell component
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- STRING:
-
Search Tool for the Retrieval of Interacting Genes/Proteins
- qRT-PCR:
-
Quantitative real-time polymerase chain reaction
- Ifi44:
-
Interferon induced protein 44
- Tnip1:
-
TNFAIP3 interacting protein 1
- Oasl1:
-
2′-5′-Oligoadenylate synthetase-like 1
- Casp4:
-
Caspase-4
- Ccl12:
-
Chemokine (C–C motif) ligand 12
- Zbp1:
-
Z-DNA-binding protein 1
- Cxcl13:
-
C-X-C motif chemokine ligand 13
- Ebi3:
-
Epstein-Barr virus–induced 3
- F10:
-
Coagulation factor X
- Traf6:
-
Flavin-containing dimethylaniline monooxygenase 3
- Stat1:
-
Signal transducer and activator of transcription 1
- JAK3:
-
Janus tyrosine kinase 3
- Rac2:
-
Rac family small GTPase 2
- Ripk1:
-
Receptor-interacting serine/threonine kinase 1
- TRAF6:
-
TNF receptor–associated factor 6
- TLRs:
-
Toll-like receptors
- Mapk3mitogen:
-
Activated protein kinase 3
- Syk:
-
Spleen tyrosine kinase
- NADPH:
-
Nicotinamide adenine dinucleotide phosphate
- GAPDH:
-
Glyceraldehyde-3-phosphate dehydrogenase
- IL-17:
-
Interleukin 17
- DC:
-
Dendritic cell
- ANOVA:
-
Analysis of variance
References
Parker, J.C. 2011. Acute lung injury and pulmonary vascular permeability: Use of transgenic models. Comprehensive Physiology. https://doi.org/10.1002/cphy.c100013.
Monahan, L.J. 2013. Acute respiratory distress syndrome. Current Problems in Pediatric and Adolescent Health Care. https://doi.org/10.1016/j.cppeds.2013.10.004.
Birkner, D.R., S. Halvachizadeh, H.C. Pape, and R. Pfeifer. 2020. Mortality of adult respiratory distress syndrome in trauma patients: A systematic review over a period of four decades. World Journal of Surgery. https://doi.org/10.1007/s00268-020-05483-5.
Liu, J., X. Wang, M. Zheng, and Q. Luan. 2018. Lipopolysaccharide from Porphyromonas gingivalis promotes autophagy of human gingival fibroblasts through the PI3K/Akt/mTOR signaling pathway. Life Sciences. https://doi.org/10.1016/j.lfs.2018.09.023.
Wang, Y., F.Y. Wang, Z. Pan, Y.Y. Dai, H.J. Xu, K.K. Jin, W.T. Wang. 2011. Effects of ambroxol combined with low-dose heparin on TNF-alpha and IL-1beta in rabbits with acute lung injury, Zhongguo Ying Yong Sheng Li Xue Za Zhi.
Li, Z., Z. Mao, Y. Lin, W. Liang, F. Jiang, J. Liu, Q. Tang, and D. Ma. 2008. Dynamic changes of tissue factor pathway inhibitor type 2 associated with IL-1β and TNF-α in the development of murine acute lung injury. Thrombosis Research. https://doi.org/10.1016/j.thromres.2008.03.019.
Qin, L., M. Li, H.L. Tan, H.X. Yang, S.D. Li, Z.X. Luan, Y.F. Chen, and M.H. Yang. 2020. Mechanistic target of rapamycin-mediated autophagy is involved in the alleviation of lipopolysaccharide-induced acute lung injury in rats. International Immunopharmacology. https://doi.org/10.1016/j.intimp.2019.105790.
Mao, K., W. Geng, Y. Liao, P. Luo, H. Zhong, P. Ma, J. Xu, Q. Tan, Y. Jin. 2020. Identification of robust genetic signatures associated with lipopolysaccharide-induced acute lung injury onset and astaxanthin therapeutic effects by integrative analysis of RNA sequencing data and GEO datasets. Aging (Albany. NY). 12,1–25.
Wang, X. 2020. Inhibition of HtrA2 alleviates inflammatory response and cell apoptosis in lipopolysaccharide-induced acute pneumonia in rats. Molecular Medicine Reports. https://doi.org/10.3892/mmr.2020.11410.
Vidal, M., M.E. Cusick, and A.L. Barabási. 2011. Interactome networks and human disease. Cell. https://doi.org/10.1016/j.cell.2011.02.016.
Liu, Y., M. Koyutürk, J.S. Barnholtz-Sloan, and M.R. Chance. 2012. Gene interaction enrichment and network analysis to identify dysregulated pathways and their interactions in complex diseases. BMC Systems Biology. https://doi.org/10.1186/1752-0509-6-65.
Xu, M., Y. Liu, Y. Huang, J. Wang, J. Yan, L. Zhang, C. Zhang. 2020. Re-exploring the core genes and modules in the human frontal cortex during chronological aging: insights from network-based analysis of transcriptomic studies. Aging (Albany. NY). https://doi.org/10.18632/aging.101589.
Altemeier, W.A., G. Matute-Bello, S.A. Gharib, R.W. Glenny, T.R. Martin, and W.C. Liles. 2005. Modulation of lipopolysaccharide-induced gene transcription and promotion of lung injury by mechanical ventilation. The Journal of Immunology. https://doi.org/10.4049/jimmunol.175.5.3369.
Britto, C.J., N. Niu, S. Khanal, L. Huleihel, J.D. Herazo-Maya, A. Thompson, M. Sauler, M.D. Slade, L. Sharma, C.S.D. Cruz, N. Kaminski, L.E. Cohn. 2019. BPIFA1 regulates lung neutrophil recruitment and interferon signaling during acute inflammation. American Journal of Physiology - Lung Cellular and Molecular Physiology. https://doi.org/10.1152/ajplung.00056.2018.
Shi, Q., R.R. Fijten, D. Spina, Y. Riffo Vasquez, V.M. Arlt, R.W. Godschalk, F.J. Van Schooten. 2017. Altered gene expression profiles in the lungs of benzo[a]pyrene-exposed mice in the presence of lipopolysaccharide-induced pulmonary inflammation. Toxicology and Applied Pharmacology. https://doi.org/10.1016/j.taap.2017.09.023.
Smith, L.S., S.A. Gharib, C.W. Frevert, and T.R. Martin. 2010. Effects of age on the synergistic interactions between lipopolysaccharide and mechanical ventilation in mice. American Journal of Respiratory Cell and Molecular Biology. https://doi.org/10.1165/rcmb.2009-0039OC.
Snyder, J.C., S.D. Reynolds, J.W. Hollingsworth, Z. Li, N. Kaminski, and B.R. Stripp. 2010. Clara cells attenuate the inflammatory response through regulation of macrophage behavior. American Journal of Respiratory Cell and Molecular Biology. https://doi.org/10.1165/rcmb.2008-0353OC.
Aggarwal, N.R., F.R. D’Alessio, K. Tsushima, V.K. Sidhaye, C. Cheadle, D.N. Grigoryev, K.C. Barnes, and L.S. King. 2010. Regulatory T cell-mediated resolution of lung injury: Identification of potential target genes via expression profiling. Physiological Genomics. https://doi.org/10.1152/physiolgenomics.00131.2009.
Zhou, G., O. Soufan, J. Ewald, R.E.W. Hancock, N. Basu, J. Xia. 2019. NetworkAnalyst 3.0: a visual analytics platform for comprehensive gene expression profiling and meta-analysis. Nucleic Acids Research. https://doi.org/10.1093/nar/gkz240.
Yu, G., L.G. Wang, Y. Han, and Q.Y. He. 2012. ClusterProfiler: An R package for comparing biological themes among gene clusters. OMICS: A Journal of Integrative Biology. https://doi.org/10.1089/omi.2011.0118.
Shi, J., Y. Zhao, Y. Wang, W. Gao, J. Ding, P. Li, L. Hu, and F. Shao. 2014. Inflammatory caspases are innate immune receptors for intracellular LPS. Nature. https://doi.org/10.1038/nature13683.
Zhao, X., Z. Yu, Z. Lv, L. Meng, J. Xu, S. Yuan, and Z. Fu. 2019. Activation of Alpha-7 Nicotinic Acetylcholine Receptors (α7nAchR) Promotes the protective autophagy in LPS-induced acute lung injury (ALI) in vitro and in vivo. Inflammation. https://doi.org/10.1007/s10753-019-01088-w.
Stenhoff, A., L. Steadman, S. Nevitt, L. Benson, R. White, B. Hill. 2020. Integrative transcriptomics analysis of lung epithelial cells and identification of repurposable drug candidates for COVID-19. Journal of Contextual Behavioral Science. 173594. https://doi.org/10.1016/j.jcbs.2020.08.008.
Wood, C., V. Kataria, and A.M. Modrykamien. 2020. The acute respiratory distress syndrome. Baylor University Medical Center Proceedings. https://doi.org/10.1080/08998280.2020.1764817.
Sapoznikov, A., Y. Gal, R. Falach, I. Sagi, S. Ehrlich, E. Lerer, A. Makovitzki, A. Aloshin, C. Kronman, T. Sabo. 2019. Early disruption of the alveolar-capillary barrier in a ricin-induced ards mouse model: neutrophil-dependent and-independent impairment of junction proteins. American Journal of Physiology - Lung Cellular and Molecular Physiology. https://doi.org/10.1152/ajplung.00300.2018.
Levitt, J.E., C.S. Calfee, B.A. Goldstein, R. Vojnik, and M.A. Matthay. 2013. Early acute lung injury: Criteria for identifying lung injury prior to the need for positive pressure ventilation. Critical Care Medicine. https://doi.org/10.1097/CCM.0b013e31828a3d99.
Yan, C., P.A. Ward, X. Wang, and H. Gao. 2013. Myeloid depletion of SOCS3 enhances LPS-induced acute lung injury through CCAAT/enhancer binding protein δ pathway. The FASEB Journal. https://doi.org/10.1096/fj.12-225797.
Wang, H.W., W. Yang, J.Y. Lu, F. Li, J.Z. Sun, W. Zhang, N.N. Guo, L. Gao, and J.R. Kang. 2013. N-acetylcysteine administration is associated with reduced activation of NF-kB and preserves lung dendritic cells function in a zymosan-induced generalized inflammation model. Journal of Clinical Immunology. https://doi.org/10.1007/s10875-012-9852-3.
Villarino, A.V., Y. Kanno, J.R. Ferdinand, and J.J. O’Shea. 2015. Mechanisms of Jak/STAT signaling in immunity and disease. The Journal of Immunology. https://doi.org/10.4049/jimmunol.1401867.
Zhu, J., Q. Yu, Y. Cai, Y. Chen, H. Liu, W. Liang, and J. Jin. 2020. Theoretical exploring selective-binding mechanisms of JAK3 by 3D-QSAR, molecular dynamics simulation and free energy calculation. Frontiers in Molecular Biosciences. https://doi.org/10.3389/fmolb.2020.00083.
Xu, W., M. Chen, N. Ge, and J. Xu. 2012. Hemagglutinin from the H5N1 virus activates Janus kinase 3 to dysregulate innate immunity. PLoS ONE. https://doi.org/10.1371/journal.pone.0031721.
Yuan, J., and Y. Zhang. 2020. Sevoflurane reduces inflammatory factor expression, increases viability and inhibits apoptosis of lung cells in acute lung injury by microRNA-34a-3p upregulation and STAT1 downregulation. Chemico-Biological Interactions. https://doi.org/10.1016/j.cbi.2020.109027.
Alimova, M., E.-H. Sidhom, A. Satyam, M. Dvela-Levitt, M. Melanson, B.T. Chamberlain, S.L. Alper, J. Santos, J. Gutierrez, A. Subramanian, E. Grinkevich, E.R. Bricio, C. Kim, A. Clark, A. Watts, R. Thompson, J. Marshall, J.L. Pablo, J. Coraor, J. Roignot, K.A. Vernon, K. Keller, A. Campbell, M. Emani, M. Racette, S. Bazua-Valenti, V. Padovano, A. Weins, S.P. McAdoo, F.W.K. Tam, L. Ronco, F. Wagner, G.C. Tsokos, J.L. Shaw, A. Greka. 2020. A high content screen for mucin-1-reducing compounds identifies fostamatinib as a candidate for rapid repurposing for acute lung injury during the COVID-19 pandemic. BioRxiv : The preprint Server for Biology. https://doi.org/10.1101/2020.06.30.180380.
Nadeem, A., S.F. Ahmad, N.O. Al-Harbi, M.M. Al-Harbi, K.E. Ibrahim, S. Kundu, S.M. Attia, W.A. Alanazi, and S.D. AlSharari. 2019. Inhibition of spleen tyrosine kinase signaling protects against acute lung injury through blockade of NADPH oxidase and IL-17A in neutrophils and γδ T cells respectively in mice. International Immunopharmacology. https://doi.org/10.1016/j.intimp.2018.12.062.
Aggarwal, N.R., L.S. King, F.R. D’Alessio. 2014. Diverse macrophage populations mediate acute lung inflammation and resolution. American Journal of Respiratory Cell and Molecular Biology. https://doi.org/10.1152/ajplung.00341.2013.
Dooley, J.L., D. Abdel-Latif, C.D. St. Laurent, L. Puttagunta, D. Befus, P. Lacy. 2009. Regulation of inflammation by Rac2 in immune complex-mediated acute lung injury. American Journal of Respiratory Cell and Molecular Biology. https://doi.org/10.1152/ajplung.90471.2008.
Waiczies, S., T. Prozorovski, C. Infante-Duarte, A. Hahner, O. Aktas, O. Ullrich, and F. Zipp. 2005. Atorvastatin induces T cell anergy via phosphorylation of ERK1. The Journal of Immunology. https://doi.org/10.4049/jimmunol.174.9.5630.
Shen, C.H., J.Y. Lin, Y.L. Chang, S.Y. Wu, C.K. Peng, C.P. Wu, and K.L. Huang. 2018. Inhibition of NKCC1 modulates alveolar fluid clearance and inflammation in ischemia-reperfusion lung injury via TRAF6-mediated pathways. Frontiers in Immunology. https://doi.org/10.3389/fimmu.2018.02049.
Najafov, A., A.K. Mookhtiar, H.S. Luu, A. Ordureau, H. Pan, P.P. Amin, Y. Li, Q. Lu, and J. Yuan. 2019. TAM kinases promote necroptosis by regulating oligomerization of MLKL. Molecular Cell. https://doi.org/10.1016/j.molcel.2019.05.022.
Wang, L., T. Wang, H. Li, Q. Liu, Z. Zhang, W. Xie, Y. Feng, T. Socorburam, G. Wu, Z. Xia, and Q. Wu. 2016. Receptor interacting protein 3-mediated necroptosis promotes lipopolysaccharide-induced inflammation and acute respiratory distress syndrome in mice. PLoS ONE. https://doi.org/10.1371/journal.pone.0155723.
Ven, V., M.A. Wemyss, J.S. Pearson. 2020. The diverse roles of RIP kinases in host-pathogen interactions. Seminars in Cell and Developmental Biology.
Qin, C., X.Y. Sai, X.F. Qian, Y. Wu, L.F. Zou, H.M. Wang, T. Bian, and Z. Yan. 2019. Close relationship between cIAP2 and human ARDS induced by severe H7N9 infection. BioMed Research International. https://doi.org/10.1155/2019/2121357.
Asad, M., A. Sabur, M. Shadab, S. Das, M. Kamran, N. Didwania, and N. Ali. 2019. EB1-3 chain of IL-35 along with TGF-β synergistically regulate anti-leishmanial immunity. Frontiers in Immunology. https://doi.org/10.3389/fimmu.2019.00616.
Genter, M.B., and S. Ali. 1998. Age-related susceptibility to 3,3’-iminodipropionitrile-induced olfactory mucosal damage. Neurobiology of Aging. https://doi.org/10.1016/S0197-4580(98)00104-3.
Ersayin, A., A. Thomas, L. Seyve, N. Thielens, M. Castellan, R. Marlu, B. Polack, and M.C. Dagher. 2017. Catalytically inactive gla-domainless factor Xa binds to TFPI and restores ex vivo coagulation in hemophilia plasma. Haematologica. https://doi.org/10.3324/haematol.2017.174037.
Funding
The research was financially supported by the National Natural Science Foundation of China (81960230, 81860241, 81960229), Basic Research Project in Guizhou Province (No.〔2019〕1354, No.〔2020〕1Y318), Basic Research Project of Zunyi (No.〔2018〕102), Young talents Found of Zunyi medical University (18zy-005).The project was supported by the National Natural Science Foundation of China (81960230, 81860241, 81960229), Basic Research Project in Guizhou Province (No.〔2019〕1354, No.〔2020〕1Y318), Basic Research Project of Zunyi (No.〔2018〕102), Young talents Found of Zunyi medical University (18zy-005).
Author information
Authors and Affiliations
Contributions
Hua Zhong and Kaimin Mao conceptualized this study research. Fang Cao performed statistical analyses. Chunyan Wang wrote the manuscript. Danling Long and Yujuan Deng helped in data consolidation and manuscript revision.
Corresponding authors
Ethics declarations
Ethics Approval and Consent to Participate
All experiment protocols conformed to the guidelines of the China Council on Animal Care and Use. These animal studies were approved by the Institutional Animal Research Committee of Union Hospital.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised: There should be an article note that will indicate that both authors’ Fang Cao and Chunyan Wang contributed equally to the paper.
SUPPLEMENTARY INFORMATION
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Cao, F., Wang, C., Long, D. et al. Network-Based Integrated Analysis of Transcriptomic Studies in Dissecting Gene Signatures for LPS-Induced Acute Lung Injury. Inflammation 44, 2486–2498 (2021). https://doi.org/10.1007/s10753-021-01518-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10753-021-01518-8