Abstract
Background
Inflammatory processes are key drivers of bronchopulmonary dysplasia (BPD), a chronic lung disease in preterm infants. In a large sample, we verify previously reported associations of genetic variants of immunology-related genes with BPD.
Methods
Preterm infants with a gestational age ≤32 weeks from PROGRESS and the German Neonatal Network (GNN) were included. Through a consensus case/control definition, 278 BPD cases and 670 controls were identified. We identified 49 immunity-related genes and 55 single-nucleotide polymorphisms (SNPs) previously associated with BPD through a comprehensive literature survey. Additionally, a quantitative genetic association analysis regarding oxygen supplements, mechanical ventilation, and continuous positive air pressure (CPAP) was performed.
Results
Five candidate SNPs were nominally associated with BPD-related phenotypes with effect directions not conflicting the original studies: rs11265269-CRP, rs1427793-NUAK1, rs2229569-SELL, rs1883617-VNN2, and rs4148913-CHST3. Four of these genes are involved in cell adhesion. Extending our analysis to all well-imputed SNPs of all candidate genes, the strongest association was rs45538638-ABCA3 with CPAP (p = 4.9 × 10−7, FDR = 0.004), an ABC transporter involved in surfactant formation.
Conclusions
Most of the previously reported associations could not be replicated. We found additional support for SNPs in CRP, NUAK1, SELL, VNN2, and ABCA3. Larger studies and meta-analyses are required to corroborate these findings.
Impact
-
Larger cohort for improved statistical power to detect genetic associations with bronchopulmonary dysplasia (BPD).
-
Most of the previously reported genetic associations with BPD could not be replicated in this larger study.
-
Among investigated immunological relevant candidate genes, additional support was found for variants in genes CRP, NUAK1, SELL, VNN2, and CHST3, four of them related to cell adhesion.
-
rs45538638 is a novel candidate SNP in reported candidate gene ABC-transporter ABCA3.
-
Results help to prioritize molecular candidate pathomechanisms in follow-up studies.
Similar content being viewed by others
Introduction
The life expectancy of preterm infants is continuously increasing due to improved clinical treatment. However, this comes at the cost of an increase in the incidence of bronchopulmonary dysplasia (BPD), especially in preterm infants with low gestational age (GA).1 For a GA of 22–25 weeks, incidence of BPD was reported to be up to 80%.2 At the current state, BPD is the most common chronic lung disease in preterm infants3 and a primary reason for their morbidity and mortality.4 Beside low GA, low birth weight (BW), especially if <1500 g, is a well-established risk factor5 for the development of BPD. The pathophysiology of the disease is the result of an extremely immature lung and is characterized by subsequent reparative processes resulting in vascular and alveolar underdevelopment6 as well as fibrotic remodeling of the lung’s anatomic structures.7 These processes can result in reduced gas exchange and hence a reduced lung function. BPD is believed to have a strong genetic background with heritability estimated from twin studies between 50 and 80%.3 Hence, six genome-wide association studies (GWASs; none resulting in genome-wide significant associations)8,9,10,11,12,13 as well as several candidate gene studies have been conducted so far (see Supplementary Table 1 for references of included associated candidate single-nucleotide polymorphisms (SNPs)). A major focus of reported candidate studies were genes involved in immune processes resulting from oxidative stress or infections promoted by the general underdeveloped state of the lung.14 These processes are believed to play an important role in the development of BPD due to their profound deleterious effects. For example, immune cells, such as macrophages or neutrophils, release oxygen radicals and other inflammatory mediators as a response to infection by bacteria or other exogenous organisms. These molecules can impair pulmonary tissue and further increase pro-inflammatory effects.1 Other immune-related genes such as surfactant proteins have multiple effects: On the one hand, they affect alveolization and capillary morphology.15,16 On the other, they are associated with immunological processes including accumulation of mast cells in the trachea and antiviral activity of the immune system.17, 18 Typically, candidate gene studies published in the past were limited by small sample sizes. Thus, replication of their findings in independent studies is required. Moreover, reported studies were typically limited regarding the number of considered candidate markers per gene. Therefore, our aim was to comprehensively evaluate previously reported immune-related candidate SNPs in a large sample of BPD cases and controls. Going beyond this replication analysis, we analyzed the respective candidate genes in more detail considering additional SNPs of these genes (see Table 1 for an overview of all the included SNPs).
Furthermore, in addition to case/control analyses we also analyzed associations with quantitative traits related to BPD, namely, duration of oxygen supplements, mechanical ventilation, and continuous positive air pressure (CPAP).
Methods
Subjects and definition of BPD
In this study, we pooled subjects from two large study groups, namely, PROGRESS (283 preterm infants recruited from 2000 to 201119 and the German Neonatal Network (GNN; 1485 preterm infants recruited from 2008 to 2010).20 Ethnic background was available from mothers only. Children from African and Asian mothers were excluded. The remaining subjects were categorized into German, non-German European (including Russian), and Turkish/Middle East genetic background of mothers. Different definitions of BPD were applied in the PROGRESS study and the GNN: The PROGRESS network included preterm infants with GA <32 weeks and excluded those showing severe malformations, administration of oligo- or anhydramnion >3 weeks before birth, or the presence of a severe metabolic disorder. In GNN, preterm infants with GA ≤36 + 6 weeks and a BW <1500 g were included. We harmonized these criteria by including only preterm infants with GA up to 32 weeks. Additionally, we included only preterm infants with high-quality genetic data, resulting in 1061 samples (for genetic filter criteria, see section “Genotyping and imputation”). Furthermore, we consistently applied a definition for mild, moderate, and severe BPD according to the definition of the National Institutes of Health21 for preterm infants in both studies. Thereby, preterm infants with supplemental oxygen ≥28 days are defined as BPD with the following degrees: Mild: no need for oxygen supplementation at 36 weeks postmenstrual age (PMA); Moderate: treatment with <30% oxygen after 36 weeks PMA; Severe: ≥30% oxygen and/or positive pressure (mechanical ventilation/CPAP) after 36 weeks PMA. In the case of deaths before 36 weeks PMA, patients with supplemental oxygen ≥28 days were considered moderate when treated with oxygen (<30%) at the time of death and severe when treated with ≥30% oxygen and/or positive pressure at the time of death. Of the 39 deceased patients, 20 deaths occurred before 36 weeks PMA and were considered moderate BPD according to our criteria. We focused on moderate and severe BPD, since heritability was estimated to be smaller for mild BPD.22 Hence, in our study, case status was defined by requirement for supplemental oxygen or mechanical ventilation after 36 weeks PMA or until death. For defining controls, we applied the following criteria: survival until 37th week PMA and no supplemental oxygen or mechanical ventilation at 36th week PMA. Using these definitions, we identified 670 controls (including 411 without BPD and 259 mild BPD patients) and 278 cases (including 140 moderate and 138 severe cases) in our studies. The remaining 113 samples of unclear classification were not considered in the analysis of case/control status but were included in quantitative trait analyses.
For the quantification of respiratory support, days of supplied oxygen, days of mechanical ventilation, and days of CPAP were considered, thereby excluding patients deceased before 36 weeks PMA. These measures were assessed as previously described.19, 20 In total, information on supplied oxygen, mechanical ventilation, and days of CPAP were available for 989 (93.2%), 1029 (97%), and 982 (92.6%) of the 1061 samples, respectively (see also Table 2 for more details and Supplementary Fig. 1 for a correlation plot). All studies were approved by local Ethics committees, and legal representatives of study subjects provided written informed consent to the study.
Genotyping and imputation
Genotypes were determined by a custom Affymetrix axiom array. Briefly, this custom array design extends the standard Axiom™ Genome-Wide CEU 1 Array design (ca 580,000 markers) by adding (a) SNPs from the genome-wide association study (GWAS) catalog,23 (b) reported expression quantitative trait loci (eQTLs) in the Chicago-eQTL-Browser,24 and (c) markers from candidate genes reported for diagnoses such as BPD, respiratory distress syndrome of the neonate, acute respiratory distress syndrome, pneumonia, sepsis, chronic obstructive pulmonary disease, and asthma in OMIM,25 PubGene (www.pubgene.com), or Phenopedia.26 Markers from genes provided by Affymetrix relevant for ADME (absorption, distribution, metabolism, and elimination), DMET (Drug Metabolism Enzymes and Transporters), and HLA (Human Leukocyte Antigen) genes were also considered. For this array design, dbSNP-Version 130 and genome build hg18 were used. A lift-over to dbSNP-Version 137 genome build hg19 was also available. The extended design finally resulted in 631,728 analyzable genetic markers. Full details of the array content are available upon request from the authors. Intensity files of successfully genotyped samples were combined and genotypes were called by Affymetrix Power Tools version 1.15 and the Axiom GT1 algorithm assuming generic priors. Samples with low call rate of autosomal SNPs (call rate <98%), samples with outlying mean squared difference of the individual’s genotype and population average genotype, sample duplicates, pairs of samples with discordant estimated and reported relatedness, and samples with discordant estimated and reported sex were excluded (excluding 187 individuals, with 1061 remaining). Reported ethnic origin was validated by principal components analysis. For this, a drop-one-in procedure was chosen including 85 European, 88 African, and 97 Asian individuals from the 1000 Genomes project (phase 1, release 3) as reference. For imputation, SNPs were selected according to the following criteria: not monomorphic, call rate ≥97%, p value of the deviation from Hardy–Weinberg equilibrium >10−6 (strata defined by mother’s country of origin27) and p value of plate association test >10−7. This resulted in 575,367 variants. Imputation was performed using 1000 Genomes phase 1 version 328 as reference (genome built hg-19/ dbSNP 135) and IMPUTE2 v2.3.029 for genotype estimation, after prephasing with SHAPEIT v2.r778.30 This resulted in 9,309,859 imputed variants with minor allele frequency (MAF) ≥0.01 and impute info score ≥0.5.
Retrieval of candidate associations from the literature
To identify appropriate candidate studies, we searched for the term “Bronchopulmonary Dysplasia” in Phenopedia26 listed in the online Human Genome Epidemiology encyclopedia. Additionally, PubMed (U.S. National Library of Medicine and the National Institutes of Health) was searched for the MeSH terms “bronchopulmonary dysplasia” and “single nucleotide polymorphisms.” Results were manually screened for candidate gene studies on BPD that considered genes with an immunological role (see Supplementary Table 1 for details). We also screened all studies cited in the identified publications. All nominally significant associations (p ≤ 0.05) with BPD or a BPD-related phenotype were considered for replication analysis in our study. We adopted the assignment of SNPs to genes as provided by the reporting study. To improve comprehensiveness, we searched PubMed again, this time using identified candidate genes of the first search round and the MeSH term “bronchopulmonary dysplasia” as search terms. Our search for candidate studies and GWASs was performed in November 2016 and updated in January 2020. Only autosomal associations were considered. This search resulted in 40 candidate studies in which 68 variants within 49 genes were reported (see Supplementary Table 1). After filtering for SNPs that could be unequivocally assigned to a biallelic dbSNP build 135 variant, 61 SNPs within 44 genes remained in analysis. Of these, 55 SNPs within 40 genes were amenable for candidate SNP replication analysis in our cohort, as they were included in the 1000 Genomes imputation reference or found in linkage disequilibrium (LD) with R2 ≥ 0.5 with a variant fulfilling our post-imputation quality-control criteria (MAF ≥0.01, info score ≥0.5). We consider support for previously reported genetic associations if at least a nominal association with a BPD-related phenotype is found in our data but exclude those with a reported effect direction that does not match the effect direction in our data. Reported genes were classified in six immunological-related subgroups (see Supplementary Table 1): extracellular processes, cellular processes, cytokines, recognition, cell adhesion, and others. The subgroup “others” comprised of genes of various immunologic subtopics not included in the former five categories. The function of the encoded protein served as the basis for this evaluation.
Going beyond direct replication of reported associations, we additionally analyzed all available variants within or near the reported 39 candidate genes. For this purpose, we considered variants fulfilling the following conditions: located within the gene or ±5000 bases 5’ or 3’ of the beginning/start of the gene (positions according to the UCSC genome browser, track UCSC genes,31 genome built hg19), MAF ≥0.01 and info score ≥0.5. In total, 8681 SNPs were included in this refined analysis (see Supplementary Tables 2 and 3 for details on SNP and gene-wide analysis).
Statistical analysis
We calculated a genetic correlation matrix by estimating pairwise relatedness from autosomal SNPs as described.32 The resulting matrix was transformed to positive semidefinite by setting negative eigenvalues to the smallest observed positive value. We used this correlation matrix to account for relatedness and ethnicity when comparing phenotypes in Table 2. We applied function “relmatGlmer” of package “lme4qtl”33 for this purpose.
Genetic associations were tested by regression modeling. We used gene doses of SNPs, i.e., assumed an additive mode of inheritance. We adjusted for sex, GA, small for gestational age (SGA, defined as BW less than the 10th percentile among infants in the same GA), and mother country. Again, analyses were adjusted for mixed ethnicities in combination with relatedness using a mixed model approach (function “polygenic” of the “GenABEL” package of R34 as previously described35). Accordingly, binary traits were analyzed as covariate and relatedness adjusted residuals of a linear model.35 Furthermore, identified nominal associations were analyzed in a BPD-trend test using the four categories: no BPD, mild, moderate, and severe BPD. This was performed again with the function “relmatLmer” of package “lme4qtl”33 with significance calculated with an approximate F-test based on the Kenward–Roger approach as implemented in the R-package pbkrtest.36 Genetic association results showed no signs of general inflation (maximum lambda was 1.01 observed for the case/control analysis). Gene-based association was carried out by applying locus scoring as implemented in FastBat37 using default parameters. Thereby, genotypes of included samples were used to calculate LD, and false discovery rates (FDRs) were calculated using the method of Benjamini and Hochberg.38 For regional association plots, LocusZoom39 was used. For comparison of observed and expected minimum p values (across all four phenotypes, namely, supplied oxygen, mechanical ventilation, CPAP, and case/control state), we compared quantiles of the observed distribution with quantiles of a beta (α = 1, β = 4) distribution. For functional annotation of SNPs, we used RegulomeDB40 and SNiPA41, which also included information on SNP deleteriousness quantified as CADD (Combined Annotation Dependent Depletion) scores.42
Results
Description of study subjects
Table 2 provides an overview of characteristics of our studied 1061 preterm infants comprising 670 controls and 278 BPD cases. The mean GA was significantly lower, the number of infants small for their GA was significantly higher, and significantly more deaths were observed in cases than in controls. The duration of supplemental oxygen, mechanical ventilation, and CPAP were also significantly higher in cases. When comparing infants with mild, moderate, and severe BPD, we observed an increasing percentage of males (53.7, 55, and 61.6%, respectively).
Association of previously reported SNPs
We analyzed 55 candidate SNPs within 40 candidate genes for which BPD associations were reported in the literature, 26 of them were directly genotyped while 29 were imputed with high quality as expressed by the imputation info score (see Supplementary Table 2). Our study was sufficiently powered to replicate the effect sizes reported in all original candidate studies and the top hits of reported GWASs. Seven nominally significant associations in seven genes were found in our study. All these hits were well imputed (info score of 1) and frequent, i.e., MAF was >5% (Table 3). Two of these associations (rs2542571-ERLEC1and rs2235587-PSMF1) showed directions inconsistent when compared with the originally reported association and therefore were not considered as supportive evidence for the originally reported association (Table 4).
The strongest association was found for the SNP rs11265269-CRP with supplemental oxygen (p = 0.005). Additional associations were found with mechanical ventilation (p = 0.025) and by the BPD-trend test (p = 0.014). C-reactive protein (CRP) is a well-known inflammatory marker and plays a major role in the body’s immune response.43 Effect direction of associations was consistent with the literature,12 with major allele T showing a protective effect. This variant is located 44 kb upstream of CRP and is according to SNiPA strongly associated with CRP levels in the UK Biobank (p = 5.35 × 10−14).
The second strongest finding was the SNP rs1427793 in NUAK1 (NUAK Family Kinase 1) associated with supplemental oxygen (p = 9.18 × 10−3). Among others, NUAK1 is involved in cell adhesion, i.e., important for relocation and movement of blood cells, including leukocytes, macrophages, and others.44 The corresponding case/control p value closely missed significance (p = 0.08). The consistency of effect directions with the literature could not be checked due to missing information in the original study. According to SNiPA, rs1327793-NUAK1 is a 3′-untranslated region variant. It was observed to affect transcription factor-binding (site ID ENSR00000436389) and histone methylation in several tissues, including the lung and blood. RegulomeDB classified this variant as class 4, i.e., weak evidence regarding binding of regulatory elements. The CADD score for this SNP was 8.7, i.e., moderately large. For this association, a stronger effect was observed in females (pinteraction = 0.006, Supplementary Fig. 2), while all other candidate variants did not show significant different effects between sexes.
Another nominally associated SNP with consistent effect direction was rs2229569-SELL (L-Selectin), a coding missense variant (proline > serine, p = 0.027 with mechanical ventilation). SELL is a cell surface protein involved in cell adhesion.45 Major Allele G was associated with less requirement of mechanical ventilation and, accordingly, shows a protective effect regarding BPD. SNiPA reported effects on transcription (cis-eQTL on SELL observed in blood, and also on SCYL3, a gene also relevant for cell adhesion). RegulomeDB evaluated the variant as class 4. Expression of SELL was reported for a variety of tissues, e.g., lung tissue and lymphocytes. High evidence for a functional consequence is indicated by its CADD score of 26.
Furthermore, variant rs1883617-VNN2 showed significant consistent association with BPD in the BPD-trend test and in case/control analysis (p = 0.027, and 0.01, respectively). When restricting controls to patients without BPD, nominal association remained (p ≤ 0.05). VNN2 is involved in transendothelial migration of neutrophils.46 The encoded gene product is a GPI-anchored cell surface molecule that takes part in transendothelial migration of neutrophils. According to SNiPA, the genetic variant is a missense coding variant, affecting amino acid sequence of VNN2. The corresponding CADD score of 16.55 is high, suggesting a functional relevance of the variant. RegulomeDB ranked it high with “1d”, i.e., evidence for regulatory effects from eQTL, transcription factor binding, motif, and DNase peak data.
Finally, we observed rs4148913-CHST3 (p = 0.037) to be nominally associated with the duration of mechanical ventilation with consistent effect direction. CHST3 is involved in cell adhesion by synthesizing chondroitin 6-sulfate47 and supposed to play a role in the maintenance of naive T lymphocytes in the spleen.48 Results on SNiPA implicated a direct regulative effect for rs4148913-CHST3 on SPOCK2. RegulomeDB reported class 4 again and the CADD score was 13.1, suggesting functional relevance of the variant.
Interestingly, 4 in these 5 genes (80%) are at least partly involved in processes related to “cell adhesion” (SELL, NUAK1, VNN2, and CHST3). This is more than expected by chance, as only13% of all analyzed genes (Supplementary Table 1) were classified as cell adhesion genes (p = 0.004, Fisher’s exact test).
Association analyses of additional variants of previously reported genes
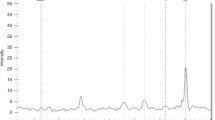
Going beyond direct replication of reported associations, we additionally analyzed all available variants within ±5000 bases 5’ or 3’ of the reported 49 candidate genes, including 9 more genes in which the variants reported in the literature could not be mapped to our genetic data. From the investigated 9469 SNPs, we observed one association withstanding multiple testing correction (see Table 5 and Fig. 1), rs45538638-ABCA3 (ATP-binding cassette sub-family A member 3). This variant was associated with CPAP (p = 4.9 × 10−7) corresponding to a (multiple testing corrected) FDR of 0.0046. The SNP is also associated with the need for supplemental oxygen (p = 0.0036), case/control status (p = 0.0078), and the BPD-trend test (p = 0.01), and shows trend association with mechanical ventilation (p = 0.06). The minor allele T is protective, i.e., associated with a decreased need for CPAP and supplemental oxygen and a decreased risk for BPD. No relevant LD with the variant rs13332514 originally reported for ABCA3 was observed because this variant is located close to a recombination hotspot of the haploblock containing rs45538638-ABCA3 (see Supplementary Fig. 3). No other SNP in LD with rs45538638-ABCA3 was included in our study, which is reasonable as rs45538638 was one of the additionally included SNPs in the framework of the custom design of our SNP array. Extending LD analysis to all variants included in 1000 Genomes phase 3 version 5 within 1 Mb, no variant in relevant LD was found either (Supplementary Table 4). RegulomeDB classified rs45538638-ABCA3 as class 2b, i.e., likely to affect binding of transcriptional factors N-Myc, Myc, and CLOCK:BMAL whose binding sites were altered by the SNP. Moreover, allele-specific chromatin modifications were predicted. Phenotypes between carriers and non-carriers of rs45538638-ABCA3-T did not differ significantly in other clinical traits like GA or SGA (Supplementary Table 5).
Gene-based locus scoring did not identify additional associations when correcting for multiple testing (Supplementary Table 6). To ensure robustness of our findings, we repeated association analyses excluding all 20 patients who died before 36 weeks PMA. In this sensitivity analysis, effect sizes were very similar to those of our main analysis and all identified associations remained statistically significant (Supplementary Table 7).
Discussion
The aim of this study was to perform a comprehensive replication analysis of reported genetic associations of BPD focusing on immunologically relevant candidate genes in a relatively large cohort of 1061 patients. We not only aimed to replicate the originally reported SNPs but also analyzed the reported genes in detail by considering other common SNPs of these genes. We were able to replicate five variant associations with effect directions not conflicting the original studies and found a new association at ABCA3.
We focus on candidate analyses rather than hypothesis-free approaches to have a reasonable power. The power of our analysis was typically >90% to replicate associations reported in previous candidate studies and the top hits of reported GWAS studies. It was also sufficient to have a decent chance of replicating suggestive candidates of GWAS studies (Table 1).
Going beyond case/control analysis, we also analyzed quantitative measures related to BPD because quantitative analyses are typically better powered. Difficulties in replicating BPD associations were also reported previously.8 Besides the above-mentioned power issue of binary traits, other explanations for this lack of direct replication are conceivable, namely, small sample sizes of reporting studies ranging from 46 to 1726, heterogeneity in the definition of case/control status across studies, publication bias of reported associations, and complexity of the phenotype, i.e., the possibility that there are no larger but many smaller genetic effects that cannot be detected with the currently available sample sizes as observed for other traits.
Of note, three of the variants associated with effect directions not conflicting the original studies were previously reported in a GWAS analysis (rs11265269-CRP,12 rs1427793-NUAK1,49 and rs4148913-CHST38) but none achieved genome-wide significance. Hence, their finding required further validation, which is achieved by our study to some extent.
However, it needs to be acknowledged that only nominal significance in replication of candidate SNPs was achieved. More robust confirmation is desirable as well as clarifying the role of the genes in the pathophysiology of BPD development.
When extending our analysis to all genotyped or imputed SNPs located in or close to the 39 previously reported candidate genes, SNP rs45538638-ABCA3 showed strong association with CPAP length withstanding stringent correction for multiple testing (p = 4.9 × 10−7, FDR = 0.004, Fig. 1). ABCA3 is a membrane-associated ATP-binding transporter of the ABC A-subfamily,50 predominantly expressed on intracellular lamellar bodies of lung alveolar type II cells of the developing and mature lung51, 52 where it is supposed to be involved in maturation of alveolar type II cells, synthesizing surfactant53 and interstitial lung fibrosis.54 A link of this gene to respiratory distress syndrome (RDS), a preliminary stage of BPD,55 was also described.56 The molecular mechanism of this association remains to be clarified. There are several transcription factors serving as candidates: binding sites of N-Myc, Myc, and CLOCK:BMAL are possibly affected by the associated variant according to RegulomeDB. Furthermore, transcription factors binding upstream can also be considered as candidates, including TTF-1 increasing gene expression of ABCA3 with relevance for lung development.57, 58 Although another study could not find any significant associations of TTF-1 variants with BPD,59 a possible connection and thus an importance of this transcription factor for the development of BPD remains. Still, the evidence of this finding does not yet allow final conclusions: on the one hand, data quality of rs45538638 was high, as it was directly genotyped on the chip. On the other hand, the MAF for allele T is only 0.022, which might cause spurious associations. Of note, in the original study which reported an association of ABCA3 with BPD,52 variant rs45538638-ABCA3 was not analyzed since these authors excluded all variants with MAF <0.1. Instead, they found an association of rs13332514-ABCA3 with requirement of supplemental oxygen at 28 postnatal days in very preterm infants (odds ratio 4.2 95% confidence interval (1.7–10.8), p = 0.002). No association was seen for this SNP in our study and rs13332514-ABCA3 showed no relevant LD with rs45538638-ABCA3 (see Supplementary Fig. 2).
One limitation of our study is that our findings cannot be readily applied to distant ethnicities, as our study design focused on probands of Caucasian descent. A second limitation is that associations with BPD may have been missed because the analysis was limited to certain candidate genes. However, given the experiences of previous GWAS for BPD and our sample size, we consider a candidate study as more appropriate to reduce the burden of multiple testing. Moreover, phenotype definitions differed between studies and standard errors are not always provided. Therefore, we chose replication analysis as our main study design. Larger studies and—as more genetic studies emerge—meta-analyses are required for a comprehensive hypothesis-free search for genetic associations of this rare phenotype, though alternative study designs such as gene-based analyses of genetically imputed expression data60 and pathway-based analyses are conceivable. In studies with sufficient sample size, a split design could also be considered. A further limitation is the adoption of SNPs to gene assignment reported in the original studies. These assignments are typically based on proximity while functional considerations would be more appropriate. Hence, we cannot exclude that observed associations might relate to other genes than the reported ones. Another limitation is that we only considered common variants. Consideration of rare variants, for example, in the framework of a gene-based analysis could add further associations.61 Finally, the definition of BPD was subject to changes over the past decades. In our study, a compromise was chosen to combine the data sets of two large study groups. A future improvement could be to implement an oxygen-challenge test to define BPD as proposed by Walsh et al.62
In summary, our results showed only limited support for previously reported candidate genes and SNP associations with BPD. Supporting evidence was found for previously reported SNP associations in or near CRP, NUAK1, SELL, VNN2, and CHST3. We also found a novel association in the ABC-transporter gene ABCA3. Further replications, GWASs of larger sample sizes, and meta-analyses are required to unravel genetics of BPD.
References
Banks-randall, B. A. & Ballard, R. A. Bronchopulmonary Dysplasia 8th edn (Elsevier, 2001).
Farstad, T., Bratlid, D., Medbø, S. & Markestad, T. Bronchopulmonary dysplasia - prevalence, severity and predictive factors in a national cohort of extremely premature infants. Acta Paediatr. 100, 53–58 (2011).
Shaw, G. M. & O’Brodovich, H. M. Progress in understanding the genetics of bronchopulmonary dysplasia. Semin. Perinatol. 37, 85–93 (2013).
Bhandari, V. et al. Familial and genetic susceptibility to major neonatal morbidities in preterm twins. Pediatrics 117, 1901–1906 (2006).
Van Marter, L. J. Epidemiology of bronchopulmonary dysplasia. Semin. Fetal Neonatal Med. 14, 358–366 (2009).
Merritt, T. A., Deming, D. D. & Boynton, B. R. The “new” bronchopulmonary dysplasia: challenges and commentary. Semin. Fetal Neonatal Med. 14, 345–357 (2009).
Hilgendorff, A. et al. Association of polymorphisms in the mannose-binding lectin gene and pulmonary morbidity in preterm infants. Genes Immun. 8, 671–677 (2007).
Wang, H. et al. A genome-wide association study (GWAS) for bronchopulmonary dysplasia. Pediatrics 132, 290–297 (2013).
Hadchouel, A. et al. Identification of SPOCK2 as a susceptibility gene for bronchopulmonary dysplasia. Am. J. Respir. Crit. Care Med. 184, 1164–1170 (2011).
Torgerson, D. G. et al. Ancestry and genetic associations with bronchopulmonary dysplasia in preterm infants. Am. J. Physiol. Cell Mol. Physiol. 315, L858–L869 (2018).
Marchini, J. & Howie, B. Genotype imputation for genome-wide association studies. Nat. Rev. Genet. 11, 499–511 (2010).
Mahlman, M. et al. Genome-wide association study of bronchopulmonary dysplasia: a potential role for variants near the CRP gene. Sci. Rep. 7, 9271 (2017).
Hamvas, A. et al. Exome sequencing identifies gene variants and networks associated with extreme respiratory outcomes following preterm birth. BMC Genet. 19, 1–10 (2018).
Saugstad, O. D. Oxygen and oxidative stress in bronchopulmonary dysplasia. J. Perinat. Med. 38, 571–577 (2010).
Coalson, J. J. Pathology of bronchopulmonary dysplasia. Semin. Perinatol. 30, 179–184 (2006).
Lahti, M., Marttila, R. & Hallman, M. Surfactant protein C gene variation in the Finnish population - association with perinatal respiratory disease. Eur. J. Hum. Genet. 12, 312–320 (2004).
Uehara, Y. et al. Surfactant protein A (SP-A) and SP-A-derived peptide attenuate chemotaxis of mast cells induced by human beta-defensin 3. Biochem. Biophys. Res. Commun. 485, 107–112 (2017).
Job, E. R. et al. Neutralizing inhibitors in the airways of naïve ferrets do not play a major role in modulating the virulence of H3 subtype influenza A viruses. Virology 494, 143–157 (2016).
Hilgendorff, A. et al. Association of polymorphisms in the human surfactant protein-D (SFTPD) gene and postnatal pulmonary adaptation in the preterm infant. Acta Paediatr. Int. J. Paediatr. 98, 112–117 (2009).
Stichtenoth, G. et al. Major contributors to hospital mortality in very-low-birth-weight infants: data of the birth year 2010 cohort of the German Neonatal Network. Klin. Padiatr. 224, 276–281 (2012).
Ehrenkranz, R. A. et al. Validation of the National Institutes of Health consensus definition of bronchopulmonary dysplasia. Pediatrics 116, 1353–1360 (2005).
Lavoie, P. M., Pham, C. & Jang, K. L. Heritability of bronchopulmonary dysplasia, defined according to the consensus statement of the National Institutes of Health. Pediatrics 122, 479–485 (2008).
Welter, D. et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 42, D1001–D1006 (2014).
Pickrell JK et al. Understanding mechanisms underlying human gene expression variation with RNA sequencing. Nature 464, 768–772 (2010).
Amberger, J. S., Bocchini, C. A., Schiettecatte, F., Scott, A. F. & Hamosh, A. OMIM.org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res. 43, D789–D798 (2015).
Yu, W., Clyne, M., Khoury, M. J. & Gwinn, M. Phenopedia and genopedia: disease-centered and gene-centered views of the evolving knowledge of human genetic associations. Bioinformatics 26, 145–146 (2009).
Troendle, J. F., & Fu, K. F. A note on testing the Hardy‐Weinberg Law. Ann. Hum. Genet. 33, 377–383 (1970).
1000 Genomes Project Consortium et al. An integrated map of genetic variation from 1,092 human genomes. Nature 491, 56–65 (2012).
Marchini, J., Howie, B., Myers, S., McVean, G. & Donnelly, P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nat. Genet. 39, 906–913 (2007).
Delaneau, O., Marchini, J. & 1000 Genomes Project Consortium. Integrating sequence and array data to create an improved 1000 Genomes Project haplotype reference panel. Nat. Commun. 5, 3934 (2014).
Lee, C. M. et al. UCSC Genome Browser enters 20th year. Nucleic Acids Res. 48, D756–D761 (2020).
Wang, J. An estimator for pairwise relatedness using molecular markers. Genetics 160, 1203–1215 (2002).
Ziyatdinov, A. et al. lme4qtl: Linear mixed models with flexible covariance structure for genetic studies of related individuals. BMC Bioinformatics 19, 1–5 (2018).
Aulchenko, Y. S., Ripke, S., Isaacs, A. & van Dujin, C. M. GenABEL: an R package for genome-wide association analysis. Bioinformatics 23, 1294–1296 (2007).
Burkhardt, R. et al. Integration of genome-wide SNP data and gene-expression profiles reveals six novel loci and regulatory mechanisms for amino acids and acylcarnitines in whole blood. PLoS Genet. 11, e1005510 (2015).
Halekoh, U. & Højsgaard, S. A kenward-Roger approximation and parametric bootstrap methods for tests in linear mixed models-the R package pbkrtest. J. Stat. Softw. 59, 1–32 (2014).
Bakshi, A. et al. Fast set-based association analysis using summary data from GWAS identifies novel gene loci for human complex traits. Sci. Rep. 6, 1–9 (2016).
Benjamini, Y. & Hochberg, Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57, 289–300 (1995).
Pruim, R. J. et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics 26, 2336–2337 (2010).
Boyle, A. P., Hong, E. L., Hariharan, M., Cheng, Y., Schaub, M. A., Kasowski, M., Karczewski, K. J., Park, J., Hitz, B. C., Weng, S., Cherry, J. M. & Snyder, M. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 22, 1790–1797 (2012).
Arnold, M., Raffler, J. Pfeufer, A. Suhre, K. & Kastenmüller, G. SNiPA: an interactive, genetic variant-centered annotation browser. Bioinformatics 31, 1334–1336 (2015).
Rentzsch, P., Witten, D., Cooper, G. M., Shendure, J. & Kircher, M. CADD: predicting the deleteriousness of variants throughout the human genome. Nucleic Acids Res. 47, D886–D894 (2019).
Gewurz, H., Mold, C., Siegel, J. & Fiedel, B. C-reactive protein and the acute phase response. Adv. Intern. Med. 27, 345–372 (1982).
Zagorska, A. et al. New roles for the LKB1-NUAK pathway in controlling myosin phosphatase complexes and cell adhesion. Sci. Signal. 3, ra25 (2010).
Carlos, B. T. M. & Harlan, J. M. Leukocyte-endothelia1 adhesion. Molecules 84, 2068–2101 (2017).
Bornhauser, B. et al. The hematopoietic stem cell marker VNN2 is associated with chemoresistance in pediatric B-cell precursor ALL. Blood Adv. 4, 4052–4064 (2020).
Liu, J. Upregulation of chondroitin 6-sulphotransferase-1 facilitates Schwann cell migration during axonal growth. J. Cell Sci. 119, 933–942 (2006).
Uchimura, K. et al. Functional analysis of the chondroitin 6-sulfotransferase gene in relation to lymphocyte subpopulations, brain development, and oversulfated chondroitin sulfates. J. Biol. Chem. 277, 1443–1450 (2002).
Ambalavanan, N. et al. Integrated genomic analyses in bronchopulmonary dysplasia. J. Pediatr. 166, 531.e13–537.e13 (2015).
Tian, W. et al. The haplotype TGGAG in the ABCA3 gene increases the risk of respiratory distress syndrome in preterm infants in Southern China. Pediatr. Neonatol. 57, 1–7 (2015).
Avery, M. E. & Mead, J. Surface properties in relation to atelectasis and hyaline membrane disease. Arch. Pediatr. Adolesc. Med. 97, 517 (1959).
Karjalainen, M. K., Haataja, R. & Hallman, M. Haplotype analysis of ABCA3: association with respiratory distress in very premature infants. Ann. Med. 40, 56–65 (2008).
Johansson, J. & Curstedt, T. Molecular structures and interactions of pulmonary surfactant components. Eur. J. Biochem. 244, 675–693 (1997).
Ota, C., Kimura, M. & Kure, S. ABCA3 mutations led to pulmonary fibrosis and emphysema with pulmonary hypertension in an 8-year-old girl. Pediatr. Pulmonol. 51, E21–3 (2016).
Ryckman, K. K., Dagle, J. M., Kelsey, K., Momany, A. M., & Murray, J. C. Genetic associations of surfactant protein D and angiotensin-converting enzyme with lung disease in preterm neonates. J Perinatol. 32, 349–355. https://doi.org/10.1038/jp.2011.104 (2012).
Anandarajan, M., Paulraj, S. & Tubman, R. ABCA3 deficiency: an unusual cause of respiratory distress in the newborn. Ulster Med. J. 78, 51–52 (2009).
Stahlman, M. T. et al. Expression of ABCA3 in developing lung and other tissues. J. Histochem. Cytochem. 55, 71–83 (2007).
Besnard, V., Xu, Y. & Whitsett, J. A. Sterol response element binding protein and thyroid transcription factor-1 (Nkx2.1) regulate Abca3 gene expression. Am. J. Physiol. Lung Cell. Mol. Physiol. 293, L1395–L1405 (2007).
Rezvani, M. et al. Association of a FGFR-4 gene polymorphism with bronchopulmonary dysplasia and neonatal respiratory distress. Dis. Markers 35, 633–640 (2013).
Barbeira, A. N. et al. Exploring the phenotypic consequences of tissue specific gene expression variation inferred from GWAS summary statistics. Nat. Commun. 9, 1825 (2018).
Li, J. et al. Exome sequencing of neonatal blood spots and the identification of genes implicated in bronchopulmonary dysplasia. Am. J. Respir. Crit. Care Med. 192, 589–596 (2015).
Walsh, M. C. Impact of a physiologic definition on bronchopulmonary dysplasia rates. Pediatrics 114, 1305–1311 (2004).
Ryckman, K. K., Dagle, J. M., Kelsey, K., Momany, A. M. & Murray, J. C. Genetic associations of surfactant protein D and angiotensin-converting enzyme with lung disease in preterm neonates. J. Perinatol. 32, 349–355 (2012).
Hadchouel, A. et al. Matrix metalloproteinase gene polymorphisms and bronchopulmonary dysplasia: identification of MMP16 as a new player in lung development. PLoS ONE 3, e3188 (2008).
Mailaparambil, B. et al. Genetic and epidemiological risk factors in the development of bronchopulmonary dysplasia. Dis. Markers 29, 1–9 (2010).
Kwinta, P., Bik-multanowski, M., Mitkowska, Z., Tomasik, T. & Legutko, M. Genetic risk factors of bronchopulmonary dysplasia. Pediatr. Res. 64, 682–688 (2008).
Pavlovic, J. et al. Genetic variants of surfactant proteins A, B, C, and D in bronchopulmonary dysplasia. Dis. Markers 22, 277–291 (2006).
Sampath, V. et al. Antioxidant response genes sequence variants and BPD susceptibility in VLBW infants. Pediatr. Res. 77, 477–483 (2015).
Cakmak, B. C. et al. Association between bronchopulmonary dysplasia and MBL2 and IL1-RN polymorphisms. Pediatr. Int. 54, 863–868 (2012).
Capoluongo, E. et al. Mannose-binding lectin polymorphisms and pulmonary outcome in premature neonates: a pilot study. Intensive Care Med. 33, 1787–1794 (2007).
Sampath, V. et al. A TLR5 (g.1174C>T) variant that encodes a stop codon (R392X) is associated with bronchopulmonary dysplasia. Pediatr. Pulmonol. 47, 460–468 (2012).
Winters, A. H. et al. Single nucleotide polymorphism in toll-like receptor 6 is associated with a decreased risk for ureaplasma respiratory tract colonization and bronchopulmonary dysplasia in preterm infants. Pediatr. Infect. Dis. J. 32, 898–904 (2013).
Bokodi, G., Derzbach, L., Banyasz, I., Tulassay, T. & Vasarhelyi, B. Association of interferon T+874A and interleukin 12 p40 promoter CTCTAA/GC polymorphism with the need for respiratory support and perinatal complications in low birthweight neonates. Arch. Dis. Child. Fetal Neonatal Ed. 92, F25–F29 (2007).
Krueger, M., Heinzmann, A., Mailaparambil, B., Härtel, C. & Göpel, W. Polymorphisms of interleukin 18 in the genetics of preterm birth and bronchopulmonary dysplasia. Arch. Dis. Child. Fetal Neonatal Ed. 96, F299–F300 (2011).
Floros, J. et al. IL-18R1 and IL-18RAP SNPs may be associated with bronchopulmonary dysplasia in African-American infants. Pediatr. Res 71, 107–114 (2012).
Prencipe, G. et al. A polymorphism in the macrophage migration inhibitory factor promoter is associated with bronchopulmonary dysplasia. Pediatr. Res. 69, 142–147 (2011).
Kukkonen, M. K. et al. Association of genes of protease-antiprotease balance pathway to lung function and emphysema subtypes. BMC Pulm. Med. 13, 36 (2013).
Kazzi, S. N. J., Kim, U. O., Quasney, M. W. & Buhimschi, I. Polymorphism of tumor necrosis factor-α and risk and severity of bronchopulmonary dysplasia among very low birth weight infants. Pediatrics 114, e243–e248 (2004).
Elhawary, N. A., Tayeb, M. T., Abdel-Ghafar, S., Rashad, M. & Alkhotani, A.-A. TNF-238 polymorphism may predict bronchopulmonary dysplasia among preterm infants in the Egyptian population. Pediatr. Pulmonol. 48, 699–706 (2013).
Manar, M. H., Brown, M. R., Gauthier, T. W. & Brown, L. A. S. Association of glutathione-S-transferase-P1 (GST-P1) polymorphisms with bronchopulmonary dysplasia. J. Perinatol. 24, 30–35 (2004).
Haas, D. M. et al. The impact of glucocorticoid polymorphisms on markers of neonatal respiratory disease after antenatal betamethasone administration. Am. J. Obstet. Gynecol. 208, 215.e1–6 (2013).
Ali, S. et al. Functional genetic variation in NFKBIA and susceptibility to childhood asthma, bronchiolitis, and bronchopulmonary dysplasia. J. Immunol. 190, 3949–3958 (2013).
Koroglu, O. A. et al. Association of vitamin D receptor gene polymorphisms and bronchopulmonary dysplasia. Pediatr. Res. 76, 171–176 (2014).
Poggi, C. et al. Genetic contributions to the development of complications in preterm newborns. PLoS ONE 10, 1–13 (2015).
Vannemreddy, P., Notarianni, C., Yanamandra, K., Napper, D. & Bocchini, J. Is an endothelial nitric oxide synthase gene mutation a risk factor in the origin of intraventricular hemorrhage? Neurosurg. Focus 28, E11 (2010).
Derzbach, L. et al. Selectin polymorphisms and perinatal morbidity in low-birthweight infants. Acta Paediatr. 95, 1213–1217 (2006).
Huusko, J. M. et al. Polymorphisms of the gene encoding Kit ligand are associated with bronchopulmonary dysplasia. Pediatr. Pulmonol. 50, 260–270 (2015).
Acknowledgements
We mourn for and express our great gratitude to Professor Ludwig Gortner, who passed away before this manuscript was completed. We greatly acknowledge his contribution to this work. Professor Gortner was Director of the Clinic for General Pediatrics and Neonatology at Saarland University Hospital and contributed samples and data from patients treated there. He continuously promoted this research and inspired us in numerous fruitful and warm-hearted discussions. We also thank the families of patients greatly for participation in the study, the PROGRESS and GNN study groups for recruitment and data collection, and Amanda Ly for copyediting.
Funding
The study was funded by the German Ministry of Education and Research in the framework of the PROGRESS study (01KI07113 and 01KI1010I: P.A., M.S.) and the German Center for Lung Research (DZL grant 82DZLJ19A2: M.S.). Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
F.B., H.K., P.A., and M.S.: substantial contributions to conception and design; P.A., T.C., M.R.T., P.N., E.-M.W., and W.G.: acquisition of data; F.B., H.K., A.G., K.H., and M.S.: analysis and interpretation of data; F.B., H.K., P.A., A.G., K.H., and M.S.: drafting the article or revising it critically for important intellectual content; F.B., H.K., P.A., T.C., A.G., K.H., M.R.T., P.N., E.-M.W., W.G., and M.S.: final approval of the version to be published.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
Consent was given by all patients according to the original networks GNN and PROGRESS.
Additional information
This study is dedicated to Professor Dr. Ludwig Gortner.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Blume, F., Kirsten, H., Ahnert, P. et al. Verification of immunology-related genetic associations in BPD supports ABCA3 and five other genes. Pediatr Res 92, 190–198 (2022). https://doi.org/10.1038/s41390-021-01689-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41390-021-01689-y