Abstract
Background
The net ammonium fluxes differ among the different root zones of Populus, but the physiological and microRNA regulatory mechanisms are unclear.
Objective
To elucidate the physiological and miRNA regulatory mechanisms, we investigated the two root zones displaying significant differences in net NH4+ effluxes of P. × canescens.
Methods
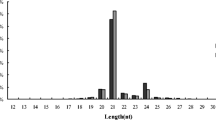
Populus plantlets were cultivated with 500 μM NH4Cl for 10 days. Six plants were randomly selected to determine the net NH4+ fluxes using a noninvasive microtest technique. High-throughput sequencing were used to determine the dynamic expression profile of miRNA among the different root zones of Populus.
Results
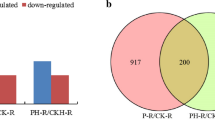
Net NH4+ efflux in zone I (from 0 to 40 mm from the root apex) was − 19.64 pmol cm−2 s−1 and in zone II (from 40 to 80 mm) it was − 43.96 pmol cm−2 s−1. The expression of eleven miRNAs was significantly upregulated, whereas fifteen miRNAs were downregulated. Moreover, eighty-eight target genes of the significantly differentially expressed miRNAs were identified in root zone II compared with zone I. Particularly, ptc-miR171a/b/e and their target, SCL6, were found to be important for the difference in net NH4+ effluxes in the two root zones. Moreover, the expression of the target of ptc-miR169d, NFYA3 was upregulated in root zone II compared with root zone I, contributing to increased NH4+ efflux and decreased NH4+ assimilation in root zone II.
Conclusion
These results indicate that miRNAs regulate the expression levels of their target genes and thus play key roles in net NH4+ fluxes and NH4+ assimilation in different poplar root zones.






Similar content being viewed by others
Availability of data
Sequence data in this article have been deposited with the NCBI GenBank Data Library under the accession codes listed in materials and methods.
References
Abarca D, Pizarro A, Hernandez I, Sanchez C, Solana SP, Del Amo A, Carneros E, Diaz-Sala C (2014) The GRAS gene family in pine: transcript expression patterns associated with the maturation-related decline of competence to form adventitious roots. BMC Plant Biol 14:354
Addo-Quaye C, Miller W, Axtell MJ (2009) CleaveLand: a pipeline for using degradome data to find cleaved small RNA targets. Bioinformatics 25:130–131
Alber A, Ehlting B, Ehlting J, Hawkins BJ, Rennenberg H (2012) Net NH4+ and NO3− flux, and expression of NH4+ and NO3− transporters in roots of Picea glauca. Trees 26:1403–1411
Cao D, Xu H, Zhao Y, Deng X, Liu Y, Soppe WJ, Lin J (2016) Transcriptome and degradome sequencing reveals dormancy mechanisms of Cunninghamia lanceolata seeds. Plant Physiol 172:2347–2362
Colmer TD, Bloom AJ (1998) A comparison of NH4+ and NO3– net fluxes along roots of rice and maize. Plant Cell Environ 21:240–246
Correa R, Baum DA (2015) Evolutionary transgenomics: prospects and challenges. Front Plant Sci 6:858
Cruz C, Lips SH, MarEins-Loução MA (1995) Uptake regions of inorganic nitrogen in roots of carob seedlings. Physiol Plant 95:167–175
Di Laurenzio L, Wysocka-Diller J, Malamy JE, Pysh L, Helariutta Y, Freshour G, Hahn MG, Feldmann KA, Benfey PN (1996) The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 86:423–433
Dluzniewska P, Gessler A, Dietrich H, Schnitzler JP, Teuber M, Rennenberg H (2007) Nitrogen uptake and metabolism in Populus × canescens as affected by salinity. New Phytol 173:279–293
Dos Santos TB, Soares JDM, Lima JE, Silva JC, Ivamoto ST, Baba VY, Souza SGH, Lorenzetti APR, Paschoal AR, Meda AR et al (2019) An integrated analysis of mRNA and sRNA transcriptional profiles in Coffea arabica L. roots: insights on nitrogen starvation responses. Funct Integr Genom 19:151–169
Fan K, Fan D, Ding Z, Su Y, Wang X (2015) Cs-miR156 is involved in the nitrogen form regulation of catechins accumulation in tea plant (Camellia sinensis L.). Plant Physiol Biochem 97:350–360
Gifford ML, Dean A, Gutierrez RA, Coruzzi GM, Birnbaum KD (2008) Cell-specific nitrogen responses mediate developmental plasticity. PNAS 105:803–808
Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ (2008) miRBase: tools for microRNA genomics. Nucleic Acids Res 36:D154-158
Gutierrez RA (2012) Systems biology for enhanced plant nitrogen nutrition. Science 336:1673–1675
Hawkins B, Boukcim H, Plassard C (2008) A comparison of ammonium, nitrate and proton net fluxes along seedling roots of Douglas-fir and lodgepole pine grown and measured with different inorganic nitrogen sources. Plant Cell Environ 31:278–287
Hawkins BJ, Robbins S, Porter RB (2014) Nitrogen uptake over entire root systems of tree seedlings. Tree Physiol 34:334–342
He XF, Fang YY, Feng L, Guo HS (2008) Characterization of conserved and novel microRNAs and their targets, including a TuMV-induced TIR-NBS-LRR class R gene-derived novel miRNA in Brassica. FEBS Lett 582:2445–2452
Huang L, Li M, Shao Y, Sun T, Li C, Ma F (2018a) Ammonium uptake increases in response to PEG-induced drought stress in Malus hupehensis Rehd. Environ Exp Bot 151:32–42
Huang L, Li M, Zhou K, Sun T, Hu L, Li C, Ma F (2018b) Uptake and metabolism of ammonium and nitrate in response to drought stress in Malus prunifolia. Plant Physiol Biochem 127:185–193
Iwamoto M, Tagiri A (2016) MicroRNA-targeted transcription factor gene RDD1 promotes nutrient ion uptake and accumulation in rice. Plant J 85:466–477
Jackson LE, Burger M, Cavagnaro TR (2008) Roots, nitrogen transformations, and ecosystem services. Plant Biol 59:341
Konishi N, Ishiyama K, Matsuoka K, Maru I, Hayakawa T, Yamaya T, Kojima S (2014) NADH-dependent glutamate synthase plays a crucial role in assimilating ammonium in the Arabidopsis root. Physiol Plant 152:138–151
Leyva-Gonzalez MA, Ibarra-Laclette E, Cruz-Ramirez A, Herrera-Estrella L (2012) Functional and transcriptome analysis reveals an acclimatization strategy for abiotic stress tolerance mediated by Arabidopsis NF-YA family members. PLoS One 7:e48138
Li H, Hu B, Wang W, Zhang Z, Liang Y, Gao X, Li P, Liu Y, Zhang L, Chu C (2016) Identification of microRNAs in rice root in response to nitrate and ammonium. J Genet Genom 43:651–661
Li S, Tian Y, Wu K, Ye Y, Yu J, Zhang J, Liu Q, Hu M, Li H, Tong Y et al (2018) Modulating plant growth-metabolism coordination for sustainable agriculture. Nature 560:595–600
Liang G, He H, Yu D (2012) Identification of nitrogen starvation-responsive microRNAs in Arabidopsis thaliana. PLoS One 7:e48951
Llave C, Xie Z, Kasschau KD, Carrington JC (2002) Cleavage of Scarecrow-like mRNA targets directed by a class of Arabidopsis miRNA. Science 297:2053–2056
Luan M, Xu M, Lu Y, Zhang Q, Zhang L, Zhang C, Fan Y, Lang Z, Wang L (2014) Family-wide survey of miR169s and NF-YAs and their expression profiles response to abiotic stress in maize roots. PLoS One 9:e91369
Luo J, Li H, Liu T, Polle A, Peng C, Luo ZB (2013a) Nitrogen metabolism of two contrasting poplar species during acclimation to limiting nitrogen availability. J Exp Bot 64:4207–4224
Luo J, Qin J, He F, Li H, Liu T, Polle A, Peng C, Luo ZB (2013b) Net fluxes of ammonium and nitrate in association with H+ fluxes in fine roots of Populus popularis. Planta 237:919–931
Marino D, Moran JF (2019) Can ammonium stress be positive for plant performance? Front Plant Sci 10:1103
Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL (2015) StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33:290–295
Pfaffl MW, Horgan GW, Dempfle L (2002) Relative expression software tool (REST©) for group-wise comparison and statistical analysis of relative expression results in real-time PCR. Nucleic Acids Res 30:e36
Qu B, He X, Wang J, Zhao Y, Teng W, Shao A, Zhao X, Ma W, Wang J, Li B et al (2015) A wheat CCAAT box-binding transcription factor increases the grain yield of wheat with less fertilizer input. Plant Physiol 167:411–423
Rewald B, Kunze ME, Godbold DL (2016) NH4: NO3 nutrition influence on biomass productivity and root respiration of poplar and willow clones. GCB Bioenergy 8:51–58
Sorin C, Declerck M, Christ A, Blein T, Ma L, Lelandais-Briere C, Njo MF, Beeckman T, Crespi M, Hartmann C (2014) A miR169 isoform regulates specific NF-YA targets and root architecture in Arabidopsis. New Phytol 202:1197–1211
Stevens ME, Woeste KE, Pijut PM (2018) Localized gene expression changes during adventitious root formation in black walnut (Juglans nigra L.). Tree Physiol 38:877–894
Sunkar R, Li YF, Jagadeeswaran G (2012) Functions of microRNAs in plant stress responses. Trends Plant Sci 17:196–203
Taylor AR, Bloom AJ (1998) Ammonium, nitrate, and proton fluxes along the maize root. Plant Cell Environ 21:1255–1263
Wang L, Mai YX, Zhang YC, Luo Q, Yang HQ (2010) MicroRNA171c-targeted SCL6-II, SCL6-III, and SCL6-IV genes regulate shoot branching in Arabidopsis. Mol Plant 3:794–806
Xu X, Yin L, Ying Q, Song H, Xue D, Lai T, Xu M, Shen B, Wang H, Shi X (2013) High-throughput sequencing and degradome analysis identify miRNAs and their targets involved in fruit senescence of Fragaria ananassa. PLoS One 8:e70959
Yu C, Chen Y, Cao Y, Chen H, Wang J, Bi YM, Tian F, Yang F, Rothstein SJ, Zhou X et al (2018a) Overexpression of miR169o, an overlapping MicroRNA in response to both nitrogen limitation and bacterial infection, promotes nitrogen use efficiency and susceptibility to bacterial blight in rice. Plant Cell Physiol 59:1234–1247
Yu H, Jiao B, Lu L, Wang P, Chen S, Liang C, Liu W (2018b) NetMiner-an ensemble pipeline for building genome-wide and high-quality gene co-expression network using massive-scale RNA-seq samples. PLoS One 13:e0192613
Zhang C, Meng S, Li Y, Zhao Z (2014) Net NH4+ and NO3− fluxes, and expression of NH4+ and NO3− transporter genes in roots of Populus simonii after acclimation to moderate salinity. Trees 28:1813–1821
Zhao Y, Wen H, Teotia S, Du Y, Zhang J, Li J, Sun H, Tang G, Peng T, Zhao Q (2017) Suppression of microRNA159 impacts multiple agronomic traits in rice (Oryza sativa L.). BMC Plant Biol 17:215
Zhong Y, Yan W, Chen J, Shangguan Z (2014) Net ammonium and nitrate fluxes in wheat roots under different environmental conditions as assessed by scanning ion-selective electrode technique. Sci Rep 4:7223
Zhou J, Liu M, Jiang J, Qiao G, Lin S, Li H, Xie L, Zhuo R (2012) Expression profile of miRNAs in Populus cathayana L. and Salix matsudana Koidz under salt stress. Mol Biol Rep 39:8645–8654
Zou N, Shi W, Hou L, Kronzucker HJ, Huang L, Gu H, Yang Q, Deng G, Yang G (2020) Superior growth, N uptake and NH4+ tolerance in the giant bamboo Phyllostachys edulis over the broad-leaved tree Castanopsis fargesii at elevated NH4+ may underlie community succession and favor the expansion of bamboo Tree. Physiolpgy 40:1606–1622
Acknowledgements
We thank Prof. Xiaohua Su and Dr. Weixi Zhang from the Research Institute of Forestry, Chinese Academy of Forestry for providing the NMT system to analyze net NH4+ fluxes.
Funding
This work was supported by the National Natural Science Foundation of China (Grant no. 31500507) and the Fundamental Research Funds for the Central Non-profit Research Institution of CAF (Grant no. CAFYBB2016QB005).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interests
The authors declare no competing interest.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhou, J., Wu, J.t. Physiological characteristics and miRNA sequencing of two root zones with contrasting ammonium assimilation patterns in Populus. Genes Genom 44, 39–51 (2022). https://doi.org/10.1007/s13258-021-01156-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-021-01156-2