Abstract
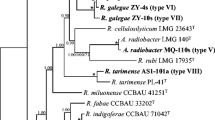
Symbiotic Paraburkholderia have been increasingly studied in the past 20 years, especially when associated with Mimosa; however, studies with native/endemic species are still scarce. In this study, thirty strains were isolated from root nodules of native Mimosa paranapiacabae and M. micropteris in two locations of the Campos Gerais. The BOX-PCR fingerprinting revealed high genomic diversity, and the 16S rRNA phylogeny clustered the strains in three distinct groups (GI, GII, GIII), with one strain occupying an isolated position. Phylogenetic analysis with four concatenated housekeeping genes (atpD + gltB + gyrB + recA) confirmed the same clusters of 16S rRNA, and the closest species were P. nodosa BR 3437T and P. guartelaensis CNPSo 3008T; this last one isolated from another Mimosa species of the Campos Gerais. The phylogenies of the symbiotic genes nodAC and nifH placed all strains in a well-supported branch with the other species of the symbiovar mimosae. The phylogenetic analyses indicated that the strains represent novel lineages of sv. mimosae and that endemic Mimosa coevolved with indigenous Paraburkholderia in their natural environments.




Similar content being viewed by others
Data availability
The following new sequences have been deposited in GenBank/EMBL/DDBJ: 16S rRNA of CNPSo 3268 (MK839064), CNPSo 3270 (MK839065), CNPSo 3272 (MK839066), CNPSo 3273 (MK839067), CNPSo 3274 (MK839068), CNPSo 3276 (MK839069), CNPSo 3278 (MK839070), CNPSo 3281 (MK839071), CNPSo 3031 (KX809741), CNPSo 3057 (KX809740), CNPSo 3058 (KX809744), CNPSo 3076 (KX809742), CNPSo 3077 (KX809743); atpD of CNPSo 3268 (MT275606), CNPSo 3270 (MT275607), CNPSo 3272 (MT275608), CNPSo 3273 (MT275609), CNPSo 3274 (MT275610), CNPSo 3276 (MT275611), CNPSo 3278 (MT275612), CNPSo 3281 (MT275613), CNPSo 3031 (MT275601), CNPSo 3057 (MT275602), CNPSo 3058 (MT275603), CNPSo 3076 (MT275604), CNPSo 3077 (MT275605); gltB of CNPSo 3268 (MT275619), CNPSo 3270 (MT275620), CNPSo 3272 (MT275621), CNPSo 3273 (MT275622), CNPSo 3274 (MT275623), CNPSo 3276 (MT275624), CNPSo 3278 (MT275625), CNPSo 3281 (MT275626), CNPSo 3031 (MT275614), CNPSo 3057 (MT275615), CNPSo 3058 (MT275616), CNPSo 3076 (MT275617), CNPSo 3077 (MT275618); gyrB of CNPSo 3268 (MK844604), CNPSo 3270 (MK844605), CNPSo 3272 (MK844606), CNPSo 3273 (MK844607), CNPSo 3274 (MK844608), CNPSo 3276 (MK844609), CNPSo 3278 (MK844610), CNPSo 3281 (MK844611), CNPSo 3031 (MK844613), CNPSo 3057 (MK844612), CNPSo 3058 (MK844616), CNPSo 3076 (MK844614), CNPSo 3077 (MK844615); recA of CNPSo 3268 (MK844617), CNPSo 3270 (MK844618), CNPSo 3272 (MK844619), CNPSo 3273 (MK844620), CNPSo 3274 (MK844621), CNPSo 3276 (MK844622), CNPSo 3278 (MK844623), CNPSo 3281 (MK844624), CNPSo 3031 (MK844626), CNPSo 3057 (MK844625), CNPSo 3058 (MK844629), CNPSo 3076 (MK844627), CNPSo 3077 (MK844628); nifH of CNPSo 3268 (MT465896), CNPSo 3270 (MT465897), CNPSo 3272 (MT465898), CNPSo 3273 (MT465899), CNPSo 3276 (MT465900), CNPSo 3278 (MT465901), CNPSo 3057 (MT465892), CNPSo 3058 (MT465893), CNPSo 3076 (MT465894), CNPSo 3077 (MT465895); nodA of CNPSo 3268 (MT465911), CNPSo 3270 (MT465912), CNPSo 3272 (MT465913), CNPSo 3273 (MT465914), CNPSo 3276 (MT465915), CNPSo 3278 (MT465916), CNPSo 3057 (MT465917), CNPSo 3058 (MT465918), CNPSo 3076 (MT465919), CNPSo 3077 (MT465920); nodC of CNPSo 3268 (MK844631), CNPSo 3270 (MK844632), CNPSo 3272 (MK844633), CNPSo 3273 (MK844634), CNPSo 3274 (MK844635), CNPSo 3276 (MK844636), CNPSo 3278 (MK844637), CNPSo 3281 (MK844638), CNPSo 3031 (KY468306), CNPSo 3057 (MK844639), CNPSo 3058 (KY468307), CNPSo 3076 (MK844640), CNPSo 3077 (MK844641).
References
Andrade ALP, Moro RS, Kuniyoshi YS, Carmo MRB (2017) Floristic survey of the Furnas Gêmeas region, Campos Gerais National Park, Paraná state, southern Brazil. Check List 13:879–899. https://doi.org/10.15560/13.6.879
Barneby RC (1991) Sensitivae censitae: a description of the genus Mimosa Linnaeus (Mimosaceae) in the new world. The New York Botanical Garden, Bronx, pp 1–5
Batista VG, Bastos RP (2014) List Anurans from a Cerrado-Atlantic Forest ecotone in Campos Gerais region, southern Brazil. Check List 10:574–582. https://doi.org/10.15560/10.3.574
Bontemps C, Elliott GN, Simon MF, dos Reis Junior FB, Gross E, Lawton RC, Neto NE, Loureiro MF, de Faria SM, Sprent JI, James EK, Young JPW (2010) Burkholderia species are ancient symbionts of legumes. Mol Ecol 19:44–52. https://doi.org/10.1111/j.1365-294X.2009.04458.x
Bontemps C, Rogel MA, Wiechmann A, Mussabekova A, Moody S, Simon MF, Moulin L, Elliott GN, Lacercat-Didier L, Dasilva C, Grether R, Camargo-Ricalde SL, Weimin C, Sprent JI, Martínez-Romero E, Young JPW, James EK (2016) Endemic Mimosa species from Mexico prefer alphaproteobacterial rhizobial symbionts. New Phytol 209:319–333. https://doi.org/10.1111/nph.13573
Bournaud C, de Faria SM, dos Santos MF, Tisseyre P, Silva M, Chaintreuil C, Gross E, James EK, Yves P, Moulin L (2013) Burkholderia species are the most common and preferred nodulating symbionts of the Piptadenia group (Tribe Mimoseae). PLoS ONE 8:e63478. https://doi.org/10.1371/journal.pone.0063478
Bournaud C, Moulin L, Cnockaert M, de Faria S, Prin Y, Severac D, Vandamme P (2017) Paraburkholderia piptadeniae sp. nov. and Paraburkholderia ribeironis sp. nov., two root-nodulating symbiotic species of Piptadenia gonoacantha in Brazil. Int J Syst Evol Microbiol 67:432–440. https://doi.org/10.1099/ijsem.0.001648
Carmo MRB, Andrade ALP, dos Santos GASD, Assis MA (2012) Structural analysis in Cerrado’s relicts at State Park of Guartelá, Tibagi, Parana State, Brazil. Ciência Florest 22:505–517. https://doi.org/10.5902/198050986618
Chen WM, Laevens S, Lee TM, Coenye T, de Vos P, Mergeay M, Vandamme P (2001) Ralstonia taiwanensis sp. nov., isolated from root nodules of Mimosa species and sputum of a cystic fibrosis patient. Int J Syst Evol Microbiol 51:1729–1735. https://doi.org/10.1099/00207713-51-5-1729
Chen WM, James EK, Coenye T, Chou J, Barrios E, de Faria SM, Elliott GN, Sheu S, Sprent JI, Vandamme P (2006) Burkholderia mimosarum sp. nov., isolated from root nodules of Mimosa spp. from Taiwan and South America. Int J Syst Evol Microbiol 56:1847–1851. https://doi.org/10.1099/ijs.0.64325-0
Chen WM, de Faria SM, James EK, Elliott GN, Lin KY, Chou JH, Sheu SY, Cnockaert M, Sprent JI, Vandamme P (2007) Burkholderia nodosa sp. nov., isolated from root nodules of the woody Brazilian legumes Mimosa bimucronata and Mimosa scabrella. Int J Syst Evol Microbiol 57:1055–1059. https://doi.org/10.1099/ijs.0.64873-0
Chibeba AM, Kyei-boahen S, Guimarães MF, Nogueira MA, Hungria M (2017) Isolation, characterization and selection of indigenous Bradyrhizobium strains with outstanding symbiotic performance to increase soybean yields in Mozambique. Agric Ecosyst Environ 246:291–305. https://doi.org/10.1016/j.agee.2017.06.017
Dall’Agnol RF, Bournaud C, de Faria SM, Béna G, Moulin L, Hungria M (2017) Genetic diversity of symbiotic Paraburkholderia species isolated from nodules of Mimosa pudica (L.) and Phaseolus vulgaris (L.) grown in soils of the Brazilian Atlantic Forest (Mata Atl antica). FEMS Microbiol Ecol 93:1–15. https://doi.org/10.1093/femsec/fix027
de Meyer SE, Cnockaert M, Ardley JK, Maker G, Yates R, Howieson JG, Vandamme P (2013) Burkholderia sprentiae sp. nov., isolated from Lebeckia ambigua root nodules. Int J Syst Evol Microbiol 63:3950–3957. https://doi.org/10.1099/ijs.0.048777-0
de Meyer SE, Briscoe L, Martínez-Hidalgo P, Agapakis CM, de-los Santos PE, Seshadri R, Reeve W, Weinstock G, O’Hara G, Howieson JG, Hirsch AM (2016) Symbiotic Burkholderia species show diverse arrangements of nif/fix and nod genes and lack typical high-affinity cytochrome cbb3 oxidase genes. Mol Plant Microbe Interact 29:609–619. https://doi.org/10.1094/MPMI-05-16-0091-R
de Oliveira ISR, Jesus EC, Ribeiro TG, da Silva MSRA, Tenorio JO, Martins LMV, de Faria SM (2019) Mimosa caesalpiniifolia Benth. adapts to rhizobia populations with differential taxonomy and symbiotic effectiveness outside of its location of origin. FEMS Microbiol Ecol 95:fiz109. https://doi.org/10.1093/femsec/fiz109
Delamuta JRM, Menna P, Ribeiro RA, Hungria M (2017) Phylogenies of symbiotic genes of Bradyrhizobium symbionts of legumes of economic and environmental importance in Brazil support the definition of the new symbiovars pachyrhizi and sojae. Syst Appl Microbiol 40:254–265. https://doi.org/10.1016/j.syapm.2017.04.005
Dobritsa AP, Samadpour M (2016) Transfer of eleven species of the genus Burkholderia to the genus Paraburkholderia and proposal of Caballeronia gen. nov. to accommodate twelve species of the genera Burkholderia and Paraburkholderia. Int J Syst Evol Microbiol 66:2836–2846. https://doi.org/10.1099/ijsem.0.001065
dos Reis Junior FB, Simon MF, Gross E, Boddey RM, Elliott GN, Neto NE, Loureiro MDF, de Queiroz LP, Scotti MR, Chen W, Nóren A, Rubio MC, de Faria SM, Bontemps C, Goi SR, Young JPW, Sprent JI, James EK (2010) Nodulation and nitrogen fixation by Mimosa spp. in the Cerrado and Caatinga biomes of Brazil. New Phytol 186:934–946. https://doi.org/10.1111/j.1469-8137.2010.03267.xKey
Dryflor (2016) Plant diversity patterns in neotropical dry forests and their conservation implications. Science 352:1383–1387. https://doi.org/10.1126/science.aaf5080
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. https://doi.org/10.1093/nar/gkh340
Elliot GN, Chen WM, Bontemps C, Chou JH, Young JPW, Sprent JI, James EK (2007) Nodulation of Cyclopia spp. (Leguminosae, Papilionoideae) by Burkholderia tuberum. Ann Bot 100:1403–1411. https://doi.org/10.1093/aob/mcm227
Estrada-de los Santos P, Palmer M, Beukes C, Steenkamp ET, Briscoe L, Id NK, Maluk M, Lafos M, Humm E, Arrabit M, Crook M, Id EG, Simon MF, dos Reis Junior FB, Whitman WB, Shapiro N, Poole PS, Hirsch AM, Venter SN, James EK (2018) Whole genome analyses suggests that Burkholderia sensu lato contains two additional novel genera (Mycetohabitans gen. nov., and Trinickia gen. nov.): implications for the evolution of diazotrophy and nodulation in the Burkholderiaceae. Genes 9:389–412. https://doi.org/10.3390/genes9080389
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791. https://doi.org/10.1111/j.1558-5646.1985.tb00420.x
Flora do Brasil (2020) Mimosa. Flora do Bras. 2020 under Constr. http://www.reflora.jbrj.gov.br/reflora/floradobrasil/FB23084 Acessed 17 Jun 2020
Garau G, Yates RJ, Deiana P, Howieson JG (2009) Novel strains of nodulating Burkholderia have a role in nitrogen fixation with papilionoid herbaceous legumes adapted to acid, infertile soils. Soil Biol Biochem 41:125–134. https://doi.org/10.1016/j.soilbio.2008.10.011
Gehlot HS, Tak N, Kaushik M, Mitra S, Chen WM, Poweleit N, Panwar D, Poonar N, Parihar R, Tak A, Sankhla IS, Ojha A, Rao SR, Simon MF, Bueno F, Perigolo N, Tripathi AK, Sprent JI, Young JPW, James EK, Gyaneshwar P (2013) An invasive Mimosa in India does not adopt the symbionts of its native relatives. Ann Bot 112:179–196. https://doi.org/10.1093/aob/mct112
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hedges SB (1992) The number of replications needed for accurate estimation of the bootstrap P value in phylogenetic studies. Mol Biol Evol 9:366–369. https://doi.org/10.1093/oxfordjournals.molbev.a040725
Helene LCF, O’Hara G, Hungria M (2020) Australia and South Africa indicates remarkable genetic diversity and reveals putative new species. Syst Appl Microbiol 43:126053. https://doi.org/10.1016/j.syapm.2020.126053
Hungria M, da Silva K (2011) Manual de curadores de germoplasma—micro-organismos: Rizóbios e bactérias promotoras do crescimento vegetal. Embrapa Recursos Genéticos e Biotecnologia; Embrapa Soja, (Documentos 333, 332), Brasília, DF
Hungria M, O’Hara GW, Zilli JE, Araujo RS, Deaker R, Howieson JG (2016) Isolation and growth of rhizobia. In: Howieson JG, Dilworth MJ (eds) Working with rhizobia. Australian Centre for International Agriculture Reserch (ACIAR), Canberra, pp 39–60
Jaccard P (1912) The distribution of flora in the alpine zone. New Phytol 11:37–50. https://doi.org/10.1111/j.1469-8137.1912.tb05611.x
Klepa MS, Urquiaga MCO, Somasegaran P, Delamuta JRM, Ribeiro RA, Hungria M (2019) Bradyrhizobium niftali sp. nov., an effective nitrogen-fixing symbiont of partridge pea [Chamaecrista fasciculata (Michx.) Greene], a native caesalpinioid legume broadly distributed in the USA. Int J Syst Evol Microbiol 69:3448–3459. https://doi.org/10.1099/ijsem.0.003640
Klonowska A, Chaintreuil C, Tiesseyre P, Miché L, Melkonian R, Ducousso M, Laguerre G, Brunel B, Moulin L (2012) Biodiversity of Mimosa pudica rhizobial symbionts (Cupriavidus taiwanensis, Rhizobium mesoamericanum) in New Caledonia and their adaptation to heavy metal-rich soils. FEMS Microbiol Ecol 81:618–635. https://doi.org/10.1111/j.1574-6941.2012.01393.x
Lammel D, Cruz LM, Carrer H, Cardoso EJBN (2013) Diversity and symbiotic effectiveness of beta-rhizobia isolated from sub-tropical legumes of a Brazilian Araucaria Forest. World J Microbiol Biotechnol 29:2335–2342. https://doi.org/10.1007/s11274-013-1400-7
Lammel DR, Cruz LM, Mescolotti D, Stürmer SL, Cardoso EJBN (2015) Woody Mimosa species are nodulated by Burkholderia in ombrophylous forest soils and their symbioses are enhanced by arbuscular mycorrhizal fungi (AMF). Plant Soil 393:123–135. https://doi.org/10.1007/s11104-015-2470-0
Lemaire B, Dlodlo O, Chimphango S, Stirton C, Schrire B, Boatwright JS, Honnay O, Smets E, Sprent J, James EK, Muasya AM (2015) Symbiotic diversity, specificity and distribution of rhizobia in native legumes of the Core Cape Subregion (South Africa). FEMS Microbiol Ecol 91:1–42. https://doi.org/10.1093/femsec/fiu024
Lemaire B, Chimphango SBM, Stirton C, Rafudeen S, Honnay O, Smets E, Chen WM, Sprent JI, James EK, Muasya AM (2016) Biogeographical patterns of legume-nodulating Burkholderia spp.: from African Fynbos to continental scales. Appl Environ Microbiol 82:5099–5115. https://doi.org/10.1128/AEM.00591-16
Lewis GP, Schrire BD, Mackinder BA, Rico L, Clark R (2013) A 2013 linear sequence of legume genera set in a phylogenetic context—a tool for collections management and taxon sampling. S Afr J Bot 89:76–84. https://doi.org/10.1016/j.sajb.2013.06.005
Liu WYY, Ridgway HJ, James TK, James EK, Chen WM, Sprent JI, Young JPW, Andrews M (2014) Burkholderia sp. induces functional nodules on the South African invasive legume Dipogon lignosus (Phaseoleae) in New Zealand soils. Microb Ecol 68:542–555. https://doi.org/10.1007/s00248-014-0427-0
LPWG (2017) A new subfamily classification of the Leguminosae based on a taxonomically comprehensive phylogeny. Taxon 66:44–77. https://doi.org/10.12705/661.3
Martínez-Romero E (2009) Coevolution in Rhizobium-legume symbiosis? DNA Cell Biol 28:361–370. https://doi.org/10.1089/dna.2009.0863
Mavima L, Beukes CW, Palmer M, de Meyer SE, James EK, Maluk M, Gross E, dos Reis Junior FB, Avontuur JR, Chan WY, Venter SN, Steenkamp ET (2020) Paraburkholderia youngii sp. nov. and “Paraburkholderia atlantica” Brazilian and Mexican Mimosa-associated rhizobia that were previously known as Paraburkholderia tuberum sv. mimosae. Syst Appl Microbiol 44:126152. https://doi.org/10.1016/j.syapm.2020.126152
Menna P, Hungria M (2011) Phylogeny of nodulation and nitrogen-fixation genes in Bradyrhizobium: supporting evidence for the theory of monophyletic origin, and spread and maintenance by both horizontal and vertical transfer. Int J Syst Evol Microbiol 61:3052–3067. https://doi.org/10.1099/ijs.0.028803-0
Mioduski J, Moro RS (2011) Functional groups of grasslands vegetation at Alagados, Ponta Grossa, Paraná. Iheringia Sér Bot 66:241–256
Mishra RPN, Tisseyre P, Melkonian R, Chaintreuil C, Miché L, Klonowska A, Gonzalez S, Bena G, Laguerre G, Moulin L (2012) Genetic diversity of Mimosa pudica rhizobial symbionts in soils of French Guiana: investigating the origin and diversity of Burkholderia phymatum and other beta-rhizobia. FEMS Microbiol Ecol 79:487–503. https://doi.org/10.1111/j.1574-6941.2011.01235.x
Moro RS, de Souza-Nogueira MKF, Milan E, Mioduski J, Pereira TK, Moro RF (2012) Grassland vegetation of Pitangui River Valley, Southern Brazil. Int J Ecosyst 2:161–170. https://doi.org/10.5923/j.ije.20120206.03
Moulin L, Muniue A, Dreyfus B (2001) Nodulation d legumes by members of the ß-subclass of Proteobacteria. Nature 411:948–950. https://doi.org/10.1038/35082070
Moulin L, James EK, Klonowska A, Faria SM, Simon MF (2015) Phylogeny, diversity, geographical distribution, and host range of legume-nodulating Betaproteobacteria: what is the role of plant taxonomy? In: de Bruijn FJ (ed) Biological nitrogen fixation. John Wiley & Sons Inc, Chichester, pp 177–190
Myers N, Mittermeier RA, Mittermeier CG, da Fonseca GAB, Kent J (2000) Biodiversity hotspots for conservation priorities. Nature 403:853–858. https://doi.org/10.1038/35002501
Nei M, Kumar S (2000) Molecular evolution and phylogenetics. Oxford University Press, Inc, New York
Oldroyd GED, Downie JA (2008) Coordinating nodule morphogenesis with Rhizobial infection in legumes. Annu Rev Plant Biol 59:519–546. https://doi.org/10.1146/annurev.arplant.59.032607.092839
Paulitsch F, Dall’Agnol RF, Delamuta JRM, Ribeiro RA, Batista JSS, Hungria M (2019a) Paraburkholderia guartelaensis sp. nov., a nitrogen-fixing species isolated from nodules of Mimosa gymnas in an ecotone considered as a hotspot of biodiversity in Brazil. Arch Microbiol 201:1435–1446. https://doi.org/10.1007/s00203-019-01714-z
Paulitsch F, Klepa MS, Silva AR, do Carmo MRB, Dall’Agnol RF, Delamuta JRM, Hungria M, Batista JSS (2019b) Phylogenetic diversity of rhizobia nodulating native Mimosa gymnas grown in a South Brazilian ecotone. Mol Biol Rep 46:529–540. https://doi.org/10.1007/s11033-018-4506-z
Paulitsch F, Dall’Agnol RF, Delamuta JRM, Ribeiro RA, Batista JSS, Hungria M (2020a) Paraburkholderia atlantica sp. nov. and Paraburkholderia franconis sp. nov., two new nitrogen-fixing nodulating species isolated from Atlantic forest soils in Brazil. Arch Microbiol 202:1369–1380. https://doi.org/10.1007/s00203-020-01843-w
Paulitsch F, Delamuta JRM, Ribeiro RA, Batista JSS, Hungria M (2020b) Phylogeny of symbiotic genes reveals symbiovars within legume-nodulating Paraburkholderia species. Syst Appl Microbiol 43:126151. https://doi.org/10.1016/j.syapm.2020.126151
Pereira-Gómez M, Ríos C, Zabaleta M, Lagurara P, Iccardi P, Azziz G, Battistoni F, Platero R (2020) Native legumes of the Farrapos protected area in Uruguay establish selective associations with rhizobia in their natural habitat. Soil Biol Biochem 148:107854. https://doi.org/10.1016/j.soilbio.2020.107854
Pires RDC, dos Reis Junior FB, Zilli JE, Fischer D, Hofmann A, James EK, Simon MF (2018) Soil characteristics determine the rhizobia in association with different species of Mimosa in central Brazil. Plant Soil 423:411–428. https://doi.org/10.1007/s11104-017-3521-5
Platero R, James EK, Rios C, Iriarte A, Sandes L, Zabaleta M, Battistoni F, Fabiano E (2016) Novel Cupriavidus strains isolated from root nodules of native Urugayan Mimosa species. Appl Environ Microbiol 82:3150–3164. https://doi.org/10.1128/AEM.04142-15.Editor
Ritter LMO, Moro RS (2007) Floristic similarities among isolated savannah plots in the higher Tibagi river catchment area, Paraná, Brazil. Terr@ Plur 1:86–98
Ritter MLO, Ribeiro MC, Moro RS (2010) Floristic composition and phytophysiognomies of Cerrado disjunct remnants in Campos Gerais, PR, Brazil—southern boundary of the biome. Biota Neotrop 10:379–412. https://doi.org/10.1590/S1676-06032010000300034
Sawana A, Adeolu M, Gupta RS (2014) Molecular signatures and phylogenomic analysis of the genus Burkholderia: proposal for division of this genus into the emended genus Burkholderia containing pathogenic organisms and a new genus Paraburkholderia gen. nov. harboring environmental species. Front Genet 5:1–22. https://doi.org/10.3389/fgene.2014.00429
Sheu S, Chou J, Bontemps C, Elliott GN, Gross E, James EK, Sprent JI, Young JPW, Chen WM (2012) Burkholderia symbiotica sp. nov., isolated from root nodules of Mimosa spp. native to north-east Brazil. Int J Syst Evol Microbiol 62:2272–2278. https://doi.org/10.1099/ijs.0.037408-0
Silva VC, Alves PAC, Rhem MFK, dos Santos JMF, James EK, Gross E (2018) Brazilian species of Calliandra Benth. (tribe Ingeae) are nodulated by diverse strains of Paraburkholderia. Syst Appl Microbiol 41:241–250. https://doi.org/10.1016/j.syapm.2017.12.003
Simon MF, Grether R, de Queiroz LP, Särkinen TE, Dutra VF, Hughes CE (2011) The evolutionary history of Mimosa (Leguminosae) towards a phylogeny of the sensitive plants. Am J Bot 98:1201–1221. https://doi.org/10.3732/ajb.1000520
Sneath P, Sokal R (1973) Numerical taxonomy: the principles and practice of numerical classification. Freeman, San Francisco
Steenkamp ET, van Zyl E, Beukes CW, Avontuur JR, Chan WY, Palmer M, Mthombeni LS, Phalane FL, Sereme TK, Venter SN (2015) Burkholderia kirstenboschensis sp. nov. nodulates papilionoid legumes indigenous to South Africa. Syst Appl Microbiol 38:545–554. https://doi.org/10.1016/j.syapm.2015.09.003
Stopnisek N, Bodenhausen N, Frey B, Fierer N, Eberl L, Weisskopf L (2014) Genus-wide acid tolerance accounts for the biogeographical distribution of soil Burkholderia populations. Environ Microbiol 16:1503–1512. https://doi.org/10.1111/1462-2920.12211
Sy A, Giraud E, Jourand P, Garcia N, Willems A, de Lajudie P, Prin Y, Neyra M, Gillis M, Boivin-Masson C, Dreyfus B (2001) Methylotrophic Methylobacterium bacteria nodulate and fix nitrogen in symbiosis with legumes. J Bacteriol 183:214–220. https://doi.org/10.1128/JB.183.1.214-220.2001
Tamashiro JY, Escobar ANG (2016) Subfamília Mimosoideae. In: Tozzi AMG, Melhem TS, Forero E, Fortuna-Perez AP, Wanderley MGL, Martins SE, Romanini RP, Pirani JR, de Melo MMF, Kizirawa M, Yano O, Cordeiro I (eds) Flora Fanerogâmica do estado de São Paulo. Instituto de Botânica, São Paulo, pp 84–166
Tamura K (1992) Estimation of the number of nucleotide substitutions when there are strong transition-transversion and G+C-content biases. Mol Biol Evol. https://doi.org/10.1093/oxfordjournals.molbev.a040752
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10:512–526
Vandamme P, Coenye T (2004) Taxonomy of the genus Cupriavidus: a tale of lost and found. Int J Syst Evol Microbiol 54:2285–2289. https://doi.org/10.1099/ijs.0.63247-0
Vandamme P, Goris J, Chen WM, de Vos P, Willems A (2002) Burkholderia tuberum sp. nov. and Burkholderia phymatum sp. nov., nodulate the roots of tropical legumes. Syst Appl Microbiol 25:507–512. https://doi.org/10.1078/07232020260517634
Versalovic J, Schneider M, Bruijn FJ, de Lupski JR (1994) Genomic fingerprinting of bacteria using repetitive sequence-based polymerase chain reaction. Methods Mol Cell Biol 5:25–40. https://doi.org/10.1128/JCM.43.1.199-207.2005
Yahara T, Javadi F, Onoda Y, de Queiroz LP, Faith DP, Prado DE, Akasaka M, Kadoya T, Ishihama F, Davies S, Slik JWF, Vatanparast M, Mimura M, Tachida H, Iwasa Y, Smith GF, Victor JE, Nkonki T (2013) Global legume diversity assessment: concepts, key indicators, and strategies. Taxon 62:249–266. https://doi.org/10.12705/622.12
Acknowledgements
M.S. Klepa and F. Paulitsch acknowledge Ph.D fellowships from CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior, Finance Code 001). V.J.B.F. Nascimento acknowledges a research fellow from Fundação Araucária. M. Hungria acknowledges a research fellow from CNPq (Brazilian Council for Scientific and Technological Development).
Funding
This study was partially supported by INCT—Plant Growth Promoting Microorganisms for Agricultural Sustainability and Environmental Responsibility/Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq 465133/2014-4), Fundação Araucária-STI 043/2019, CAPES and Fundação Araucária (09/2016).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no ethical conflicts.
Ethics approval and consent to participate
The authors declare that they have consented to participate in the manuscript and publish it.
Additional information
Communicated by Erko Stackebrandt.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Klepa, M.S., Janoni, V., Paulitsch, F. et al. Molecular diversity of rhizobia-nodulating native Mimosa of Brazilian protected areas. Arch Microbiol 203, 5533–5545 (2021). https://doi.org/10.1007/s00203-021-02537-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00203-021-02537-7