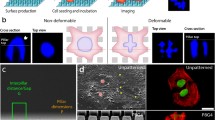
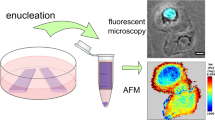
Abstract
Nuclear mechanics is emerging as a key component of stem cell function and differentiation. While changes in nuclear structure can be visually imaged with confocal microscopy, mechanical characterization of the nucleus and its sub-cellular components require specialized testing equipment. A computational model permitting cell-specific mechanical information directly from confocal and atomic force microscopy of cell nuclei would be of great value. Here, we developed a computational framework for generating finite element models of isolated cell nuclei from multiple confocal microscopy scans and simple atomic force microscopy (AFM) tests. Confocal imaging stacks of isolated mesenchymal stem cells were converted into finite element models and siRNA-mediated Lamin A/C depletion isolated chromatin and Lamin A/C structures. Using AFM-measured experimental stiffness values, a set of conversion factors were determined for both chromatin and Lamin A/C to map the voxel intensity of the original images to the element stiffness, allowing the prediction of nuclear stiffness in an additional set of other nuclei. The developed computational framework will identify the contribution of a multitude of sub-nuclear structures and predict global nuclear stiffness of multiple nuclei based on simple nuclear isolation protocols, confocal images and AFM tests.







Similar content being viewed by others
Availability of data and material
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
Code availability
The code generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Andalib MN, Lee JS, Ha L, Dzenis Y, Lim JY (2016) Focal adhesion kinase regulation in stem cell alignment and spreading on nanofibers. Biochem Biophys Res Commun 473:920–925. https://doi.org/10.1016/j.bbrc.2016.03.151
Bas G et al (2020) Low intensity vibrations augment mesenchymal stem cell proliferation and differentiation capacity during in vitro expansion. Sci Rep 10:9369. https://doi.org/10.1038/s41598-020-66055-0
Cao G, Chandra N (2010) Evaluation of biological cell properties using dynamic indentation measurement. Phys Rev E 81:021924. https://doi.org/10.1103/PhysRevE.81.021924
Cao G, Sui J, Sun S (2013) Evaluating the nucleus effect on the dynamic indentation behavior of cells. Biomech Model Mechanobiol 12:55–66. https://doi.org/10.1007/s10237-012-0381-z
Darling EM, Di Carlo D (2015) High-throughput assessment of cellular mechanical properties. Annu Rev Biomed Eng 17:35–62. https://doi.org/10.1146/annurev-bioeng-071114-040545
Dupont S et al (2011) Role of YAP/TAZ in mechanotransduction. Nature 474:179–183
Engler AJ, Sen S, Sweeney HL, Discher DE (2006) Matrix elasticity directs stem cell lineage specification. Cell 126:677–689. https://doi.org/10.1016/j.cell.2006.06.044
Ghosh S et al (2019) Deformation microscopy for dynamic intracellular and intranuclear mapping of mechanics with high spatiotemporal resolution. Cell Rep 27:1607-1620.e1604. https://doi.org/10.1016/j.celrep.2019.04.009
Guilluy C et al (2014) Isolated nuclei adapt to force and reveal a mechanotransduction pathway in the nucleus. Nat Cell Biol 16:376–381. https://doi.org/10.1038/ncb2927
Hanna H, Mir LM, Andre FM (2018) In vitro osteoblastic differentiation of mesenchymal stem cells generates cell layers with distinct properties. Stem Cell Res Ther 9:203. https://doi.org/10.1186/s13287-018-0942-x
Harris AR, Jreij P, Fletcher DA (2016) Mechanotransduction by the actin cytoskeleton: converting mechanical stimuli into biochemical signals. Annu Rev Biophys 47:617–631. https://doi.org/10.1146/annurev-biophys-070816-033547
Heo SJ et al (2015) Biophysical regulation of chromatin architecture instills a mechanical memory in mesenchymal stem cells. Sci Rep 5:16895. https://doi.org/10.1038/srep16895
Hoffman LM et al. (2020) Mechanical stress triggers nuclear remodeling and the formation of Transmembrane Actin Nuclear lines with associated Nuclear Pore Complexes. Mol biol cell, mbc.E19-01-0027, https://doi.org/10.1091/mbc.E19-01-0027
Humphrey JD, Dufresne ER, Schwartz MA (2014) Mechanotransduction and extracellular matrix homeostasis. Nat Rev Mol Cell Biol 15:802–812. https://doi.org/10.1038/nrm3896
Lammerding J et al (2006) Lamins A and C but not lamin B1 regulate nuclear mechanics. J Biol Chem 281:25768–25780. https://doi.org/10.1074/jbc.M513511200
Le HQ et al (2016) Mechanical regulation of transcription controls Polycomb-mediated gene silencing during lineage commitment. Nat Cell Biol 18:864–875. https://doi.org/10.1038/ncb3387
Makhija E, Jokhun DS, Shivashankar GV (2016) Nuclear deformability and telomere dynamics are regulated by cell geometric constraints. Proc Natl Acad Sci USA 113:E32-40. https://doi.org/10.1073/pnas.1513189113
Martins RP, Finan JD, Farshid G, Lee DA (2012) Mechanical regulation of nuclear structure and function. Annu Rev Biomed Eng 14:431–455. https://doi.org/10.1146/annurev-bioeng-071910-124638
McGarry JG, Prendergast PJ (2004) A three-dimensional finite element model of an adherent eukaryotic cell. Eur Cells Mater 7:27–33; discussion 33–24
Nagayama K, Yamazaki S, Yahiro Y, Matsumoto T (2014) Estimation of the mechanical connection between apical stress fibers and the nucleus in vascular smooth muscle cells cultured on a substrate. J Biomech. https://doi.org/10.1016/j.jbiomech.2014.01.042
Newberg J et al (2020) Isolated nuclei stiffen in response to low intensity vibration. J Biomech. https://doi.org/10.1016/j.jbiomech.2020.110012
Niu T, Cao G (2014) Finite size effect does not depend on the loading history in soft matter indentation. J Phys D Appl Phys 47:385303. https://doi.org/10.1088/0022-3727/47/38/385303
Pagnotti GM et al (2019) Combating osteoporosis and obesity with exercise: leveraging cell mechanosensitivity. Nat Rev Endocrinol. https://doi.org/10.1038/s41574-019-0170-1
Peister A et al (2004) Adult stem cells from bone marrow (MSCs) isolated from different strains of inbred mice vary in surface epitopes, rates of proliferation, and differentiation potential. Blood 103:1662–1668. https://doi.org/10.1182/blood-2003-09-3070
Rubin J, Styner M, Uzer G (2018) Physical signals may affect mesenchymal stem cell differentiation via epigenetic controls. Exerc Sport Sci Rev 46:42–47. https://doi.org/10.1249/jes.0000000000000129
Sankaran JS et al (2020) Knockdown of formin mDia2 alters lamin B1 levels and increases osteogenesis in stem cells. Stem Cells 38:102–117. https://doi.org/10.1002/stem.3098
Shiu J-Y, Aires L, Lin Z, Vogel V (2018) Nanopillar force measurements reveal actin-cap-mediated YAP mechanotransduction. Nat Cell Biol 20:262–271. https://doi.org/10.1038/s41556-017-0030-y
Stephens AD, Banigan EJ, Marko JF (2018a) Separate roles for chromatin and lamins in nuclear mechanics. Nucleus (austin, Tex) 9:119–124. https://doi.org/10.1080/19491034.2017.1414118
Stephens AD et al (2018b) Chromatin histone modifications and rigidity affect nuclear morphology independent of lamins. Mol Biol Cell 29:220–233. https://doi.org/10.1091/mbc.E17-06-0410
Tang G, Galluzzi M, Zhang B, Shen YL, Stadler FJ (2019) Biomechanical heterogeneity of living cells: comparison between atomic force microscopy and finite element simulation. Langmuir: ACS J Surf Colloids 35:7578–7587. https://doi.org/10.1021/acs.langmuir.8b02211
Wang L, Wang L, Xu L, Chen W (2019) Finite element modelling of single cell based on atomic force microscope indentation method. Comput Math Methods Med 2019:7895061. https://doi.org/10.1155/2019/7895061
Acknowledgements
We greatly appreciate the AFM expertise from Dr. Paul Davis and the Surface Sciences Laboratory.
Funding
NASA ISGC NNX15AI04H, NIH awards R01AG059923, 5P2CHD086843-03, P20GM109095, P20GM103408 and NSF awards 1929188 & 2025505 .
Author information
Authors and Affiliations
Contributions
ZK was involved in the data analysis/interpretation, manuscript writing and final approval of manuscript. JN contributed to the data analysis/interpretation and data analysis. MG contributed to the data analysis/interpretation, data analysis and final approval of manuscript. SJ was involved in the data analysis/interpretation, manuscript writing and final approval of manuscript. CKF contributed to the financial support, data analysis/interpretation and final approval of manuscript. GU contributed to the concept/design, financial support, data analysis/interpretation, manuscript writing and final approval of manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The author(s) declare no competing interests financial or otherwise.
Ethical approval
All methods were carried out in accordance with relevant guidelines and regulations of Boise Institutional Animal Care and Use Committee and Institutional Biosafety Committee. All procedures were approved by Boise State University Institutional Animal Care and Use Committee, and Institutional Biosafety Committee.
Consent for publication
All authors consent to publication.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kennedy, ., Newberg, ., Goelzer, . et al. Modeling stem cell nucleus mechanics using confocal microscopy. Biomech Model Mechanobiol 20, 2361–2372 (2021). https://doi.org/10.1007/s10237-021-01513-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10237-021-01513-w