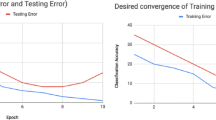
Abstract
Compressed sensing magnetic resonance imaging (CS-MRI) has been a widely studied field in biomedical signal processing owing to its success and practical MR machines integrated with the technology already rolling on in market. Since then, a decade-long research in the field has paved its way into deep-learning techniques being incorporated into the field to simplify some of the complex, theoretical issues that the original setup pose, without compromising efficiency. These machine-learning methods are particularly useful in context of input signal adaptability. The data acquisition process of MRI is noisy in nature with various types of noises associated, such as Rician noise, Gaussian Noise, and motion artifacts like breathing artifacts. However, the repercussions of noise has been scarcely incorporated into the study of most of these methods. In this context, training a model directly with the obtained data is somewhat inefficient. This paper proposes a Generative Adversarial Network (GAN)-based technique for obtaining a concise and more representative model which attempts to be a more robust noise immune network. Keeping in mind increased efficiency and reduced reconstruction time, the proposed method attempts to address the problem of MR image reconstruction. The results obtained from the proposed method has been compared with traditional CS-MRI and other contemporary methods using objective parameters such as PSNR, SSIM index, BRISQUE and FID score, and subjective parameters such as mean opinion score and LPIPS.








Similar content being viewed by others
References
M. Sandilya, S.R. Nirmala, Compressed sensing trends in magnetic resonance imaging. Eng. Sci. Technol. Int. J. 20(4), 1342–1352 (2017)
M. Lustig, D. Donoho, J.M. Pauly, Sparse MRI: The application of compressed sensing for rapid MR imaging. Magn. Reson. Med. Off. J. Int. Soc. Magn. Reson. Med. 58(6), 1182–1195 (2007)
S. Ravishankar, Y. Bresler, MR image reconstruction from highly undersampled k-space data by dictionary learning. IEEE Trans. Med. Imaging 30(5), 1028–1041 (2010)
Y. Huang, J. Paisley, Q. Lin, X. Ding, X. Fu, X.-P. Zhang, Bayesian nonparametric dictionary learning for compressed sensing MRI. IEEE Trans. Image Process. 23(12), 5007–5019 (2014)
M. Aharon, M. Elad, A. Bruckstein, K-SVD: an algorithm for designing overcomplete dictionaries for sparse representation. IEEE Trans. Signal Process. 54(11), 4311–4322 (2006)
J. Schlemper, G. Yang, P. Ferreira, A. Scott, L.-A. McGill, Z. Khalique, M. Gorodezky, M. Roehl, J. Keegan, D. Pennell et al., Stochastic Deep Compressive Sensing for the Reconstruction of Diffusion Tensor Cardiac Mri (Springer, Berlin, 2018), pp. 295–303
I. Goodfellow, J. Pouget-Abadie, M. Mirza, B. Xu, D. Warde-Farley, S. Ozair, A. Courville, Y. Bengio, Generative Adversarial Nets. Adv. Neural Inf. Process. Syst. 1, 2672–2680 (2014)
T.M. Quan, T. Nguyen-Duc, W.-K. Jeong, Compressed sensing MRI reconstruction using a Generative Adversarial Network with a cyclic loss. IEEE Trans. Med. Imaging 37(6), 1488–1497 (2018)
K. Lei, M. Mardani, J.M. Pauly, S.S. Vasanawala, Wasserstein GANs for MR imaging: from paired to unpaired training. IEEE Trans. Med. Imaging 40, 105–115 (2020)
M. Manimala, C.D. Naidu, M.G. Prasad, Sparse MR image reconstruction considering Rician noise models: a CNN approach. Wirel. Pers. Commun. 116(1), 491–511 (2021)
G. Yang, S. Yu, H. Dong, G. Slabaugh, P.L. Dragotti, X. Ye, F. Liu, S. Arridge, J. Keegan, Y. Guo et al., DAGAN: deep de-aliasing generative adversarial networks for fast compressed sensing MRI reconstruction. IEEE Trans. Med. Imaging 37(6), 1310–1321 (2017)
R. Shaul, I. David, O. Shitrit, T.R. Raviv, Subsampled brain mri reconstruction by generative adversarial neural networks. Med. Image Anal. 65, 101747 (2020)
J. Lv, J. Zhu, G. Yang, Which GAN? a comparative study of generative adversarial network-based fast MRI reconstruction. Philos. Trans. R. Soc. A 379(2200), 20200203 (2021)
Q. Lyu, C. You, H. Shan, and G. Wang, Super-resolution MRI through deep learning. arXiv:1810.06776 (2018)
Q. Lyu, C. You, H. Shan, Y. Zhang, G. Wang, Super-resolution MRI and CT through GAN-CIRCLE. Proc. SPIE 11113, Developments in X-Ray Tomography XII, 111130X (2019)
Y. Chen, F. Shi, A.G. Christodoulou, Y. Xie, Z. Zhou, D. Li, In Efficient and Accurate MRI Super-Resolution Using a Generative Adversarial Network and 3D Multi-Level Densely Connected Network (Springer, Berlin, 2018), pp. 91–99
Q. Yang, P. Yan, Y. Zhang, H. Yu, Y. Shi, X. Mou, M.K. Kalra, Y. Zhang, L. Sun, G. Wang, Low-dose CT image denoising using a generative adversarial network with Wasserstein distance and perceptual loss. IEEE Trans. Med. Imaging 37(6), 1348–1357 (2018)
P. Deora, B. Vasudeva, S. Bhattacharya, P.M. Pradhan, Structure preserving compressive sensing MRI reconstruction using generative adversarial networks. in Proceedings of the IEEE Computer Society Conference on Computer Vision and Pattern Recognition Workshops, pp. 2211–2219, Seattle, WA, USA (2020)
M. Cheon, J.-H. Kim, J.-H. Choi, J.-S. Lee, Generative adversarial network-based image super-resolution using perceptual content losses. in ECCV Workshop (2018)
W.-S. Lai, J.-B. Huang, N. Ahuja, M.-H. Yang, Fast and accurate image super-resolution with deep laplacian pyramid networks. IEEE Trans. Pattern Anal. Mach. Intell. 41(11), 2599–2613 (2018)
H. Gudbjartsson, S. Patz, The rician distribution of noisy MRI data. Magn. Reson. Med. 34(6), 910–914 (1995)
K. Zhang, W. Zuo, Y. Chen, D. Meng, L. Zhang, Beyond a gaussian denoiser: residual learning of deep CNN for image denoising. IEEE Trans. Image Process. 26(7), 3142–3155 (2017)
W. Jifara, F. Jiang, S. Rho, M. Cheng, S. Liu, Medical image denoising using convolutional neural network: a residual learning approach. J. Supercomput. 75(2), 704–718 (2019)
X. You, N. Cao, H. Lu, M. Mao, W. Wanga, Denoising of MR images with Rician noise using a wider neural network and noise range division. Magn. Reson. Imaging 64, 154–159 (2019)
M. Bouhrara, D.A. Reiter, H. Celik, J.-M. Bonny, V. Lukas, K.W. Fishbein, R.G. Spencer, Incorporation of Rician noise in the analysis of biexponential transverse relaxation in cartilage using a multiple gradient echo sequence at 3 and 7 tesla. Magn. Reson. Med. 73(1), 352–366 (2015)
J.V. Manjón, J. Carbonell-Caballero, J.J. Lull, G. García-Martí, L. Martí-Bonmatí, M. Robles, MRI denoising using non-local means. Med. Image Anal. 12(4), 514–523 (2008)
M. Arjovsky, S. Chintala, L. Bottou, Wasserstein generative adversarial networks. in International conference on machine learning, pp. 214–223 (2017)
L. Weng, From GAN to WGAN. arXiv:1904.08994 (2019)
I. Gulrajani, F. Ahmed, M. Arjovsky, V. Dumoulin, A.C. Courville, Improved training of Wasserstein GANs. Adv. Neural Inform. Process. Syst. 5767–5777 (2017)
J. Adler, S. Lunz, Banach Wasserstein GAN. Adv. Neural Inf. Process. Syst. 6754–6763 (2018)
P. Coupé, P. Yger, S. Prima, P. Hellier, C. Kervrann, C. Barillot, An optimized blockwise nonlocal means denoising filter for 3-D Magnetic Resonance images. IEEE Trans. Med. Imaging 27(4), 425–441 (2008)
Z. Yuan, M. Jiang, Y. Wang, B. Wei, Y. Li, P. Wang, W. Menpes-Smith, Z. Niu, G. Yang, SARA-GAN: self-attention and relative average discriminator based generative adversarial networks for fast compressed sensing MRI reconstruction. Front. Neuroinformatics 14, 1 (2020)
Y. Guo, C. Wang, H. Zhang, G. Yang, Deep Attentive Wasserstein Generative Adversarial Networks for MRI Reconstruction with Recurrent Context-Awareness (Springer, Berlin, 2020), pp. 167–177
S. Yu, H. Dong, G. Yang, G. Slabaugh, P.L. Dragotti, X. Ye, F. Liu, S. Arridge, J. Keegan, D. Firmin, et al., Deep de-aliasing for fast compressive sensing mri. arXiv:1705.07137 (2017)
J. Lv, C. Wang, G. Yang, Pic-gan: a parallel imaging coupled generative adversarial network for accelerated multi-channel mri reconstruction. Diagnostics 11(1), 61 (2021)
H. Zhao, O. Gallo, I. Frosio, J. Kautz, Loss functions for image restoration with neural networks. IEEE Trans. Comput. Imaging 3(1), 47–57 (2016)
Y. Blau, R. Mechrez, R. Timofte, T. Michaeli, L. Zelnik-Manor, The 2018 pirm challenge on perceptual image super-resolution. arXiv:1809.07517 (2018)
J. Deng, W. Dong, R. Socher, L. Li, K. Li, L. Fei-Fei, ImageNet: a large-scale hierarchical image database. IEEE Conference on Computer Vision and Pattern Recognition, pp. 248-255 (2009). https://doi.org/10.1109/CVPR.2009.5206848.
R. Zhang, P. Isola, A.A. Efros, E. Shechtman, O. Wang, The unreasonable effectiveness of deep features as a perceptual metric. in Proc. CVPR 2018, pp. –586–595
A. Hore, D. Ziou, Image quality metrics: PSNR vs. SSIM, 20 International Conference on Pattern Recognition, pp. 2366-2369 (2010)
A. Mittal, A.K. Moorthy, A.C. Bovik, No-reference image quality assessment in the spatial domain. IEEE Trans. Image Process. 21(12), 4695–4708 (2012)
M. Sandilya, S. Nirmala, Determination of reconstruction parameters in compressed sensing MRI using BRISQUE score. IEEE Conference on Information, Communication, Engineering and Technology(ICICET), 1–5 (2018)
M. Heusel, H. Ramsauer, T. Unterthiner, B. Nessler, S. Hochreiter, Gans trained by a two time-scale update rule converge to a local nash equilibrium. arXiv:1706.08500 2017
M. Seitzer, pytorch-fid: FID Score for PyTorch. https://github.com/mseitzer/pytorch-fid, Version 0.1.1 (2020)
A. Krizhevsky, I. Sutskever, G.E. Hinton, Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Process. Syst. 25, 1097–1105 (2012)
F. Knoll, T. Murrell, A. Sriram, N. Yakubova, J. Zbontar, M. Rabbat, A. Defazio, M.J. Muckley, D.K. Sodickson, C.L. Zitnick et al., Advancing machine learning for MR image reconstruction with an open competition: overview of the 2019 fastMRI challenge. Magn. Reson. Med. 84, 3054–3070 (2020)
W. Huang, X. Li, Y. Chen, X. Li, M.-C. Chang, M.J. Oborski, D.I. Malyarenko, M. Muzi, G.H. Jajamovich, A. Fedorov et al., Variations of dynamic contrast-enhanced magnetic resonance imaging in evaluation of breast cancer therapy response: a multicenter data analysis challenge. Transl. Oncol 7(1), 153–166 (2014)
C.A. Cocosco, V. Kollokian, R.K.-S. Kwan, G.B. Pike, A.C. Evans, Brainweb: Online interface to a 3d MRI simulated brain database. NeuroImage (1997)
L. Marak, J. Cousty, L. Najman, H. Talbot, 4d Morphological Segmentation and the MICCAI LV-Segmentation Grand Challenge, MICCAI 2009 Workshop on Cardiac MR Left Ventricle Segmentation Challenge, vol. 1 (MIDAS, USA, 2009), pp. 1–8
C. McCollough, TU-FG-207A-04: overview of the low dose CT grand challenge. Med. Phys. 43(6Part35), 3759–3760 (2016)
Acknowledgements
The authors would like to thank Center for High Performance Computing, a collaboration of Center for Development of Advanced computing (C-DAC) and Assam Engineering College, Guwahati, Assam, India, for allowing access to the laboratory to perform necessary research.
Funding
Self-funded
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
None.
Availability of Data and Material
Dataset—publicly available FastMRI dataset, url - https://fastmri.med.nyu.edu/
Code Availability
The following packages/repositories have been made use of or been customized in the manuscript: PIX2PIX - https://github.com/phillipi/pix2pix Keras WGAN -https://github.com/keras-team/keras-contrib/blob/master/examples/improved_wgan.py liveplotloss - https://github.com/stared/livelossplot VGG -16 -https://gist.github.com/baraldilorenzo/07d7802847aaad0a35d3 SPORCO - https://github.com/bwohlberg/sporco
Ethics Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
I give my consent for the publication of the following manuscript, which can include photograph(s) and/or details within the text (“Material”) to be published in the Journal—Applied Magnetic Resonance. I have discussed this consent statement with S.R. Nirmala and Navajit Saikia, who are authors of this paper. I understand that this copy may be available in both print and on the Internet, and will be available to a broader audience through marketing channels and other third parties. Therefore, anyone can read material published in the Journal.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sandilya, M., Nirmala, S.R. & Saikia, N. Compressed Sensing MRI Reconstruction Using Generative Adversarial Network with Rician De-noising. Appl Magn Reson 52, 1635–1656 (2021). https://doi.org/10.1007/s00723-021-01416-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00723-021-01416-0