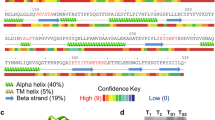
Abstract
We have identified and cloned five auxin conjugate amidohydrolases (M20D peptidases) in four different hornwort species (Phaeoceros carolinianus, Megaceros tosanus, Megaceros vincentianus, and Paraphymatoceros hallii). Sequence analysis suggests that all five enzymes have greater than 60% overall similarity to tracheophyte amidohydrolases. Phylogenetic analysis supports the hypothesis that the bryophyte and tracheophyte hydrolases are derived from a common ancestor. Enzyme studies of hornwort auxin amidohydrolases all demonstrate greater activity and substrate recognition than the more ancient liverwort hydrolase (MpILR1). The mean wild-type hornwort hydrolytic activity (23.2 ± 6.8 pmol auxin released/min/ml), although higher than the liverwort activity (1.3 ± 1.1 pmol auxin released/min/ml), was almost a magnitude lower than the average activity in tracheophyte hydrolases (186.2 ± 52.1 pmol auxin released/min/ml). Two hornwort orthologues from M. vincentianus and M. tosanus possess a Glycine238/240 replacing the tracheophytically conserved Serine209, while two from P. hallii and P. carolinianus have an Alanine238 at that homologous residue location. Further enzymatic studies and three-dimensional structural analyses of the hornwort enzymes present supporting evidence that the Ala238-line of hornworts is the likely clade from which tracheophytes arose.





Similar content being viewed by others
Change history
12 September 2021
A Correction to this paper has been published: https://doi.org/10.1007/s00344-021-10479-z
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215(3):403–410
Bandurski RS, Cohen JD, Slovin JP, Reinecke DM (1995) Auxin biosynthesis and metabolism. In: Davies PJ (ed) Plant Hormones, 2nd edn. Kluwer Academic Publishers, Boston, Massachusetts, pp 39–65
Bitto E, Bingman CA, Bittova L, Houston NL, Boston RS, Fox BG, Philips GN (2009) X-ray structure of ILL2, an auxin-conjugate amidohydrolase from Arabidopsis thaliana. Proteins 74:61–71
Blakeslee JJ, Spatola Rossi T, Kriechbaumer V (2019) Auxin biosynthesis: spatial regulation and adaptation to stress. J of Expt Bot 70(19):5041–5049
Bradford M (1976) A rapid and sensitive method for the quantification of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72(1–2):248–254
Campanella JJ, Ludwig-Müller J, Town CD (1996) Isolation and characterization of mutants of Arabidopsis thaliana with increased resistance to growth inhibition by indoleacetic acid amino acid conjugates. Plant Physiol 112:735–745
Campanella JJ, Larko D, Smalley J (2003a) A molecular phylogenomic analysis of the ILR1-like family of IAA amidohydrolase genes. Comp Funct Genomics 4:584–600
Campanella JJ, Bitincka L, Smalley JV (2003b) MatGAT: An application that generates similarity/identity matrices using protein or DNA sequences. BMC Bioinformatics 4:29
Campanella JJ, Olajide A, Magnus V, Ludwig-Müller J (2004) A novel auxin conjugate from wheat with substrate specificity for longer side-chain auxin amide conjugates. Plant Physiol 135:2230–2240
Campanella JJ, Smith SM, Leibu D, Wexler S, Ludwig-Müller J (2008) The auxin conjugate hydrolase family of Medicago truncatula and their expression during the interaction with two symbionts. J Plant Growth Reg 27(1):26–38
Campanella JJ, Zaben N, Enriquez D, Smalley JV, Ludwig-Mueller J (2014) An enzymatic analysis of Loblolly pine and Sitka spruce auxin conjugate hydrolases and evolutionary implications. Acta Hort 1042:79–88
Campanella JJ, Kurdach S, Bochis J, Smalley JV (2018) Evidence for exaptation of the Marchantia polymorpha M20D peptidase MpILR1 into the tracheophyte auxin regulatory pathway. Plant Physiol 177(4):1595–1604
Campanella JJ, Kurdach S, Skibitski R, Smalley JV, Desind S, Ludwig-Müller J (2019) Evidence for the early evolutionary loss of the M20D auxin amidohydrolase family from mosses and horizontal gene transfer from soil bacteria of cryptic hydrolase orthologous to Physcomitrella patens. J Plant Growth Reg 38:1428–1438
Casanova-Sáez R, Mateo-Bonmatí E, Ljung K (2021) Auxin metabolism in plants. Cold Spring Harb Perspect Biol 13:a039867
Chou JC, Welch WH, Cohen J (2004) His-404 and His-405 are essential for enzyme catalytic activities of a bacterial indole-3-acetyl-l-aspartic acid hydrolase. Plant Cell Physiol 45(9):1335–1341
Cooke TJ, Poli DB, Sztein AE, Cohen JD (2002) Evolutionary patterns in auxin action. Plant Mol Biol 49:319–338
Cooke TJ, Poli D, Cohen JD (2003) Did auxin play a crucial role in the evolution of novel body plans during the Late Silurian-Early Devonian radiation of land plants? In: Hemsly AM, Poole I (eds) The Evolution of Plant Physiology. Academic Press London, U.K., pp 85–107
Davies RT, Goetz GH, Lasswell J, Anderson MN, Bartel B (1999) IAR3 encodes an auxin conjugate hydrolase from Arabidopsis. Plant Cell 11(3):365–376
Eisenberg D, Weiss RM, Terwilliger TC, Wilco W (1982) Hydrophobic moments and protein structure. Faraday Symp Chem Soc 17:109–120
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Gower JC (2015) Principal Coordinates Analysis. Wiley StatsRef: Statistics Reference Online, pp 1–7
Harris BJ, Harrison CJ, Hetherington AM, Williams TA (2020) Phylogenomic evidence for the monophyly of bryophytes and the reductive evolution of stomata. Current Biol 30(11):2001-2012.e2
Hori K, Maruyama F, Fujisawa T, Togashi T, Yamamoto N, Seo M, Sato S, Yamada T, Mori H, Tajima N (2014) Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nat Commun 5:3978–3986
LeClere S, Tellez R, Rampey RA, Matsuda SPT, Bartel B (2002) Characterization of a family of IAA-amino acid conjugate hydrolases from Arabidopsis. J Biol Chem 277:20446–20452
Ligrone R, Duckett JG, Renzaglia KS (2012) Major Transitions in the evolution of early land plants: a bryological perspective. Ann Bot 109:851–871
Ljung K, Hull AK, Kowalczyk M, Marchant A, Celenza J, Cohen JD, Sandberg G (2002) Biosynthesis, conjugation, catabolism and homeostasis of indole-3-acetic acid in Arabidopsis thaliana. Plant Mol Biol 49:249–272
Lu H, Zhao WM, Zheng Y (2005) Analysis of synonymous codon usage bias in Chlamydia. Acta Biochim Biophys Sin (Shanghai) 37(1):1–10
Ludwig-Müller J (2011) Auxin conjugates: their role for plant development and in the evolution of land plants. J Exp Bot 62:1757–1773
Ludwig-Müller J, Epstein E, Hilgenberg W (1996) Auxin-conjugate hydrolysis in Chinese cabbage: characterization of an amidohydrolase and its role during infection with clubroot disease. Physiol Plant 97(4):627–634
Ludwig-Müller J, Decker EL, Reski R (2009) Dead end for auxin conjugates in Physcomitrella? Plant Signal Behav 4:116–118
Normanly J, Bartel B (1999) Redundancy as a way of life—IAA metabolism. Curr Opin Plant Biol 2:207–213
Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’Hara RB, Simpson GL, Solymos P (2016) Vegan: Community Ecology Package. R package version 2.4–1
Page RD (1996) TREEVIEW: an application to display phylogenetic trees on personal computers. Comput Appl Biosci 12:357–358
Poli DB, Jacobs M, Cooke TJ (2003) Auxin regulation of axial growth in bryophyte sporophytes: its potential significance for the evolution of early land plants. Am J Bot 90(10):1405–1415
Puttick MN, Morris JL, Williams TA, Cox CJ, Edwards D, Kenrick P, Pressel S, Wellman CH, Schneider H, Pisani D, Donoghue PCJ (2018) The interrelationships of land plants and the nature of the ancestral embryophyte. Curr Biol 28:733–745
R Core Team (2014) R: A language and environment for statistical computing. R Foundation for Statistical Computing. Vienna, Austria
Rawlings ND, Barrett AJ (1995) Evolutionary families of metallopeptidases. Methods Enzymol 248:183–228
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York
Sokoloff DD, Remizowa MV (2020) Diversity, development and evolution of archegonia in land plants. Bot J Linn Soc 20:1–40
Suzuki H, Kohich T, Nishihama R (2021) Auxin biology in bryophyta: a simple platform with versatile functions. Cold Spring Harb Perspect Biol 13:a040055
Sztein AE, Cohen JD, García de la Fuente I, Cooke TJ (1999) Auxin metabolism in mosses and liverworts. Am J Bot 86:1544–1555
Sztein AE, Cohen JD, Cooke TJ (2000) Evolutionary patterns in the auxin metabolism of green plants. Int J Plant Sci 161:849–859
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24:4872–4882
Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, Lepore R, Schwede T (2018) SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46(W1):W296–W303
Wickham H (2009) ggplot2: Elegant Graphics for Data Analysis. Springer Publishing Company, New York, New York
Wong GKS, Soltis DE, Leebens-Mack J, Wickett NJ, Barker MS, Peer YV, Graham SW, Melkonian M (2020) Sequencing and analyzing the transcriptomes of a thousand species across the tree of life for green plants. Annu Rev Plant Biol 71:741–765
Zhang J, Fu XX, Li RQ, Xiang Zhao X, Yang Liu Y, Li M-H, Zwaenepoel A, Hong Ma H (2020) The hornwort genome and early land plant evolution. Nat Plants 6:107–118
Acknowledgements
We thank Jerry Cohen for his donation of several auxin conjugates that are no longer commercially available. We would also like to thank Lisa Campanella for her help in editing this manuscript and thank you Scott Kight for your common sense. This work was supported by a Margaret and Herman Sokol Fellow Award (#MHSFA07022), as well as a Wehner Foundation Grant (#WFG19001).
Author information
Authors and Affiliations
Contributions
J.J.C. conceived the project, performed phylogenetic analysis, 3D protein modeling, analyzed data, cloned and characterized PcILR2A238S, and wrote the article with the contributions of all the authors; E.A.M. cloned MvILR1, MtIL1, and PhILR1 and characterized them; S.D. cloned PcILR1 and PcILR2 and characterized them; A.H. designed and cloned MtILRG240S and MvILRG238S and characterized them; A.A. designed and cloned PhILRA238S and characterized it; J.V.S. performed the codon usage analysis and helped to analyze data.
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Additional information
Handling Editor: Stephen Pollmann.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this article was revised: “The Supplementary Table 1 was originally published with the wrong format; it has now been replaced with the correct file”.
Supplementary Information
Below is the link to the electronic supplementary material.
344_2021_10467_MOESM1_ESM.jpg
Supplementary file1 Principal coordinate analysis of codon usage to determine if hornwort hydrolases were bacterial contaminants acquired during genomic sequencing. The two-dimensional plot was created using ggplot2 (Wickham 2009) (jpg 86 kb)
344_2021_10467_MOESM2_ESM.jpg
Supplementary file2 Amino acid sequence similarity matrix of ILR1 orthologues from various Plantae species and several prokaryotes. The matrix was generated with MatGAT v1.1 using the default values for protein analysis. The color variation (green to red) indicates levels of similarity: green-shaded values have a higher similarity, whereas red-shaded values have a lower similarity (jpg 397 kb)
344_2021_10467_MOESM3_ESM.jpg
Supplementary file3 Amino acid alignment of orthologue amidohydrolases. Alignment was performed using CLUSTAL X v2.1. Red Letters indicate conserved amino acids (Cys137, His139, Glu173, His197, and His397). The red arrow indicates where the homologous Ser209 can be located. The blue bands designate the sequence of the hornwort amidohydrolases (jpg 736 kb)
344_2021_10467_MOESM4_ESM.jpg
Supplementary file4 3D-modelling analysis of the protein structures of A) AtILL2, B) PcILR1, C) PcILR2, D) PhILR, E) MtILR, and F) MvILR. The red amino acids, indicated by arrows, are conserved and found in the enzymes’ active site. The green bar indicates the alignment between the residue homologues of Ser209 (in AtILL2) and the conserved Phenylalanine across from it (Phe381 in AtILL2) (jpg 552 kb)
344_2021_10467_MOESM5_ESM.jpg
Supplementary file5 3D-modelling analysis of A) PcILR1 and B) PcILR2 examining further polymorphic structural differences (AA76, AA208, AA221). Close-up view of active-site around C) PcILR1 Ser221 and D) PcILR2 Gly221 to examine topologic changes. Red arrows indicate residues which are conserved between the two proteins. Black arrows indicate amino acid polymorphisms (jpg 405 kb)
Rights and permissions
About this article
Cite this article
Medina, E.A., Desind, S., Hallak, A. et al. Examination of the M20D Auxin Conjugate Peptidase Family from Hornwort and Implications on the Evolution of the Tracheophytes. J Plant Growth Regul 41, 2695–2706 (2022). https://doi.org/10.1007/s00344-021-10467-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00344-021-10467-3