Abstract
Fluorene is a harmful organic toxicant extensively disseminated in the water and dry land ecosystem. Its toxicity and ubiquitous presence pose issues concerning its biodegradation. Characterization of the molecular mechanisms of fluorene degradation, detection of metabolites, and appraisal of its viability in toxicant removal by the SMT-1 Pseudomonas sp. strain are the main purposes of this study. In this work, the catabolic intermediates were identified from resting cell reactions of the SMT-1 strain as well as the involved catabolic pathway of fluorene. Based on liquid chromatography mass spectrometry analysis, the identified intermediates were 9-fluorenone; 3,4-dihydroxy-9-fluorenone; phthalate and protocatechuic acid. The specific primers were designed to amplify the fluorene-degrading 4921 dioxygenase gene segment from the SMT-1 Pseudomonas sp. strain. The 4921 dioxygenase gene was expressed, purified and characterized. The apparent Km and Vmax values were 25.99 µM min−1 and 0.77 U mg−1, respectively. The enzyme was most active at pH 7.5 and 25 °C in Tris–HCl buffer and was identified by measuring the initial reaction velocity for 1 min. Effect of metal salts on enzyme activity was accessed to see the impact on protein stability. Most of the analyzed metal salts inhibited enzyme activity to different degrees, and exhibited very low activity in the presence of FeCl3. Understanding the physiological, metabolic pathway and molecular mechanism of fluorene degradation is an important factor in increasing significant information of this biological process. This strain may serve as a potential candidate for further use in the bioremediation process to treat organic toxicant contaminated sites.



Similar content being viewed by others
References
Boldrin B, Tiehm A, Fritzsche C (1993) Degradation of phenanthrene, fluorene, fluoranthene, and pyrene by a Mycobacterium sp. Appl Environ Microbiol 59:1927–1930
Casellas M, Grifoll M, Bayona J, Solanas A (1997) New metabolites in the degradation of fluorene by Arthrobacter sp. strain F101. Appl Environ Microbiol 63:819–826
Cerniglia E, Sutherland J, Cro WS (1992) Fungal metabolism of aromatic hydrocarbons. In: Winkelmann G (Ed) Microbial degradation of natural products, VCH Verlagsegesellschaft, Weinheim
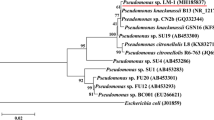
Desta M, Wang W, Zhang L, Ping X, Tang H (2019) Isolation, characterization and genomic analysis of pseudomonas sp. strain SMT-1, an efficient fluorene-degrading bacterium. J Evolut Bioinf 15:1–7
Grifoll M, Selifonov S, Chapman P (1994) Evidence for a novel pathway in the degradation of fluorine by Pseudomonas sp. strain F274. Appl Environ Microbiol 60:2438–2449
Grifoll M, Selifonov S, Gatlin C, Chapman P (1995) Actions of a versatile fluorene-degrading bacterial isolate on polycyclic aromatic hydrocarbons. Appl Environ Microbiol 61:3711–3723
Hadibarata T, Teh ZC (2014) Optimization of pyrene degradation by white-rot fungus Pleurotus pulmonarius F043 and characterization of its metabolites. Bioprocess Biosyst Eng 37(8):1679–1684
Kawasaki S, Jin F, Takada T (2011) High dispersion power of cardo-typed fluorene. INTECH Open Access Publisher, Moieties on Carbon Fillers
Kumar S, Stiches G, Tamura K (2016) MEGA 7: molecular genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Latimer J, Zheng J (2003) The sources, transport and fate of PAHin the marine environment. In: Douben PET (Ed) PAHs an ecotoxicological perspective, John Wiley, New York
Liu Y, Wang L, Huang K, Wang W, Nie X, Jiang Y, Li P, Liu S, Xu PHZ (2014) Physiological and biochemical characterization of a novel Nicotine-degrading Bacterium Pseudomonas geniculata N1. PLoS ONE 9:399
Monna L, Omori T, Kodama T (1993) Microbial degradation of dibenzofuran, fluorene, and dibenzo-p-dioxin by Staphylococcus auriculans DBF63. Appl Environ Microbiol 59:285–289
Mrozik A, Piotrowska-Seget Z, Labuzek S, Polish J (2013) Environ. Studies 12:15–25
Sayed K, Baloo L, Sharma NK (2021) Bioremediation of total petroleum hydrocarbons (TPH) by bioaugmentation and biostimulationin water with floating oil spill containment booms as bioreactor basin. Int J Environ Res Public Health 18:2226
Souza HML, Taniguchi S, Bícego MC (2015) Polycyclic aromatic hydrocarbons in superficial sediments of the Negro River in the Amazon region of Brazil. Jbraz Chem Soc 26:1438–1449
Syed JH, Iqbal M, Zhong G (2017) Polycyclic Aromatic hydrocarbons in Chinese Forest soils: profile composition, spatial variations and source apportionment. Sci Rep 7:2692
Trenz S, Engesser K, Fischer P, Knackmuss H (1994) Degradation of fluorene by Brevibacterium sp. strain DPO1361: a novel C-C bond cleavage mechanism via 1, 10-dihydro-1, 10-dihydroxyfluorene-9-one. J Bacteriol 176:789–795
Veeranagouda Y, Paul PE, Gorla P, Siddavattam D, Karegoudar TB (2006) Complete mineralization of dimethylformamide by Ochrobactrum sp. DGVK1 isolated from the soil samples collected from the coalmine left overs. Appl Microbiol Biotechnol 71:369–375
Wattiau P, Bastiaens L, van Herwijnen R (2011) Fluorene degradation by Sphin-gomonas sp. LB126 proceeds through protocatechuic acid: a genetic analysis. Res Microbiol 152:861–872
Acknowledgements
This work was supported by grants from National Key Research and Development Project (SQ2018YFA090024), the Science and Technology Commission of Shanghai Municipality (17JC1403300), by the “Shuguang Program” (17SG09) supported by Shanghai Education Development Foundation and Shanghai Municipal Education Commission, and by grants from the Chinese National Science Foundation for Excellent Young Scholars (31422004).
Author information
Authors and Affiliations
Contributions
HZT outset and designed experiments. MD, ZLG and WW performed experiments. HZT and PX contributed reagents and materials. MD performed all the experiment and data analyses and wrote the manuscript. All authors discussed and revised the manuscript. All authors commented on the manuscript before submission. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare they have no competing interests.
Accession numbers
The Pseudomonas sp. strain SMT-1 has been deposited in the China Center for Type Culture Collection (CCTCC) under accession number AB2018208. The whole-genome shotgun sequencing data have been deposited at DDBJ/ENA/Gen Bank under accession number QJOV00000000. The version described in this article is the first version QJOV01000000.
Rights and permissions
About this article
Cite this article
Desta, M., Zhang, L., Wang, W. et al. Molecular mechanisms and biochemical analysis of fluorene degradation by the Pseudomonas sp. SMT-1 strain. 3 Biotech 11, 416 (2021). https://doi.org/10.1007/s13205-021-02946-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-021-02946-x