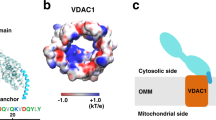
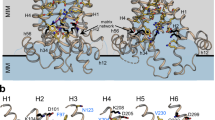
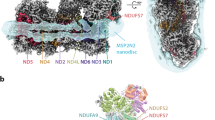
Abstract
The import of thiamine pyrophosphate (TPP) through both mitochondrial membranes was studied using a total of 3-µs molecular dynamics simulations. Regarding the translocation through the mitochondrial outer membrane, our simulations support the conjecture that TPP uses the voltage-dependent anion channel, the major pore of this membrane, for its passage to the intermembrane space, as its transport presents significant analogies with that used by other metabolites previously studied, in particular with ATP. As far as passing through the mitochondrial inner membrane is concerned, our simulations show that the specific carrier of TPP has a single binding site that becomes accessible, through an alternating access mechanism. The preference of this transporter for TPP can be rationalized mainly by three residues located in the binding site that differ from those identified in the ATP/ADP carrier, the most studied member of the mitochondrial carrier family. The simulated transport mechanism of TPP highlights the essential role, at the energetic level, of the contributions coming from the formation and breakage of two networks of salt bridges, one on the side of the matrix and the other on the side of the intermembrane space, as well as the interactions, mainly of an ionic nature, formed by TPP upon its binding. The energy contribution provided by the cytosolic network establishes a lower barrier than that of the matrix network, which can be explained by the lower interaction energy of TPP on the matrix side or possibly a uniport activity.










Similar content being viewed by others
Abbreviations
- AAC:
-
ADP/ATP carrier
- c-state:
-
Cytoplasmic-open state
- ffTK:
-
Force field tool kit
- IMS:
-
Intermembrane space
- m-state:
-
Matrix-open state
- MIM:
-
Mitochondrial inner membrane
- MOM:
-
Mitochondrial outer membrane
- MCF:
-
Mitochondrial carrier family
- TM:
-
Transmembrane
- TPP:
-
Thiamine pyrophosphate
- TPPT:
-
Thiamine pyrophosphate transporter
- VDAC:
-
Voltage dependent anion channel
References
Bettendorff L (1994) The compartmentation of phosphorylated thiamine derivatives in cultured neuroblastoma cells. Biochim Biophys Acta Mol Cell Res 1222:7–14. https://doi.org/10.1016/0167-4889(94)90019-1
Bettendorff L, Wins P (2009) Thiamin diphosphate in biological chemistry: new aspects of thiamin metabolism, especially triphosphate derivatives acting other than as cofactors. FEBS J 276:2917–2925. https://doi.org/10.1111/j.1742-4658.2009.07019.x
Kale S, Ulas G, Song J, Brudvig GW, Furey W, Jordan F, Kale S (2008) Efficient coupling of catalysis and dynamics in the E1 component of Escherichia coli pyruvate dehydrogenase multienzyme complex. Proc Natl Acad Sci USA 105:1158–1163. https://doi.org/10.1073/pnas.0709328105
Rosenberg MJ, Agarwala R, Bouffard G, Davis J, Fiermonte G, Hilliard MS, Koch T, Kalikin LM, Makalowska I, Morton DH, Petty EM, Weber JL, Palmieri F, Kelley RI, Schäffer AA, Biesecker LG (2002) Mutant deoxynucleotide carrier is associated with congenital microcephaly. Nat Genet 32:175–179. https://doi.org/10.1038/ng948
Dolce V, Fiermonte G, Runswick MJ, Palmieri F, Walker JE (2000) The human mitochondrial deoxynucleotide carrier and its role in the toxicity of nucleoside antivirals. Proc Natl Acad Sci USA 98:2284–2288. https://doi.org/10.1073/pnas.031430998
Lindhurst MJ, Fiermonte G, Song S, Struys E, De Leonardis F, Schwartzberg PL, Chen A, Castegna A, Verhoeven N, Mathews CK, Palmieri F, Biesecker LG (2006) Knockout of Slc25a19 causes mitochondrial thiamine pyrophosphate depletion, embryonic lethality, CNS malformations, and anemia. Proc Natl Acad Sci 103:15927–15932. https://doi.org/10.1073/pnas.0607661103
Rostovtseva TK, Komarov A, Bezrukov SM, Colombini M (2002) Dynamics of nucleotides in VDAC channels: structure-specific noise generation. Biophys J 82:193–205. https://doi.org/10.1016/S0006-3495(02)75386-1
Mannella CA, Bonner WD (1975) Biochemical characteristics of the outer membranes of plant mitochondria. BBA Biomembr 413:213–225. https://doi.org/10.1016/0005-2736(75)90105-4
Krammer E-M, Vu GT, Homblé F, Prévost M (2015) Dual mechanism of ion permeation through VDAC revealed with inorganic phosphate ions and phosphate metabolites. PLoS ONE 10:e0121746. https://doi.org/10.1371/journal.pone.0121746
Benz R (1994) Permeation of hydrophilic solutes through mitochondrial outer membranes: review on mitochondrial porins. Biochim Biophys Acta 1197:167–196
Homblé F, Krammer E-M, Prévost M (1818) Plant VDAC: facts and speculations. Biochim Biophys Acta 2012:1486–1501. https://doi.org/10.1016/j.bbamem.2011.11.028
Colombini M (1989) Voltage gating in the mitochondrial channel, VDAC. J Membr Biol 111:103–111. https://doi.org/10.1007/BF01871775
Bayrhuber M, Meins T, Habeck M, Becker S, Giller K, Villinger S, Vonrhein C, Griesinger C, Zweckstetter M, Zeth K (2008) Structure of the human voltage-dependent anion channel. Proc Natl Acad Sci USA 105:15370–15375
Hiller S, Garces RG, Malia TJ, Orekhov VY, Colombini M, Wagner G (2008) Solution structure of the integral human membrane protein VDAC-1 in detergent micelles. Science 321:1206–1210. https://doi.org/10.1126/science.1161302
Ujwal R, Cascio D, Colletier J-P, Faham S, Zhang J, Toro L, Ping P, Abramson J (2008) The crystal structure of mouse VDAC1 at 2.3 A resolution reveals mechanistic insights into metabolite gating. Proc Natl Acad Sci 105:17742–17747. https://doi.org/10.1073/pnas.0809634105
Choudhary OP, Paz A, Adelman JL, Colletier J-P, Abramson J, Grabe M (2014) Structure-guided simulations illuminate the mechanism of ATP transport through VDAC1. Nat Struct Mol Biol 21:626–632. https://doi.org/10.1038/nsmb.2841
Schredelseker J, Paz A, López CJ, Altenbach C, Leung CS, Drexler MK, Chen J-N, Hubbell WL, Abramson J (2014) High-resolution structure and double electron-electron resonance of the Zebrafish voltage dependent anion channel 2 reveal an oligomeric population. J Biol Chem 289:12566–12577. https://doi.org/10.1074/jbc.M113.497438
Hosaka T, Okazaki M, Kimura-Someya T, Ishizuka-Katsura Y, Ito K, Yokoyama S, Dodo K, Sodeoka M, Shirouzu M (2017) Crystal structural characterization reveals novel oligomeric interactions of human voltage-dependent anion channel 1. Protein Sci 26:1749–1758. https://doi.org/10.1002/pro.3211
Böhm R, Amodeo GF, Murlidaran S, Chavali S, Wagner G, Winterhalter M, Brannigan G, Hiller S (2020) The structural basis for low conductance in the membrane protein VDAC upon β-NADH binding and voltage gating. Structure 28:206-214.e4. https://doi.org/10.1016/j.str.2019.11.015
Colombini M (2009) The published 3D structure of the VDAC channel: native or not? Trends Biochem Sci 34:382–389. https://doi.org/10.1016/j.tibs.2009.05.001
Colombini M (1863) The VDAC channel: molecular basis for selectivity. Biochim Biophys Acta 2016:2498–2502. https://doi.org/10.1016/j.bbamcr.2016.01.019
Hiller S, Abramson J, Mannella C, Wagner G, Zeth K (2010) The 3D structures of VDAC represent a native conformation. Trends Biochem Sci 35:514–521. https://doi.org/10.1016/j.tibs.2010.03.005
Krammer E-M, Homblé F, Prévost M (2011) Concentration dependent ion selectivity in VDAC: a molecular dynamics simulation study. PLoS ONE 6:e27994. https://doi.org/10.1371/journal.pone.0027994
Mlayeh L, Krammer E-M, Léonetti M, Prévost M, Homblé F (1858) The mitochondrial VDAC of bean seeds recruits phosphatidylethanolamine lipids for its proper functioning. Biochim Biophys Acta Bioenerg 2017:786–794. https://doi.org/10.1016/j.bbabio.2017.06.005
Van Liefferinge F, Krammer EM, Sengupta D, Prévost M (2019) Lipid composition and salt concentration as regulatory factors of the anion selectivity of VDAC studied by coarse-grained molecular dynamics simulations. Chem Phys Lipids 220:66–76. https://doi.org/10.1016/j.chemphyslip.2018.11.002
Krammer E-M, Homblé F, Prévost M (1828) Molecular origin of VDAC selectivity towards inorganic ions: a combined molecular and Brownian dynamics study. Biochim Biophys Acta 2013:1284–1292. https://doi.org/10.1016/j.bbamem.2012.12.018
Krammer E-M, Saidani H, Prévost M, Homblé F (2014) Origin of ion selectivity in Phaseolus coccineus mitochondrial VDAC. Mitochondrion 19:206–213. https://doi.org/10.1016/j.mito.2014.04.003
Noskov SY, Rostovtseva TK, Bezrukov SM (2013) ATP transport through VDAC and the VDAC-tubulin complex probed by equilibrium and nonequilibrium MD simulations. Biochemistry 52:1–3. https://doi.org/10.1021/bi4011495
Rui H, Il Lee K, Pastor RW, Im W (2011) Molecular dynamics studies of ion permeation in VDAC. Biophys J 100:602–610. https://doi.org/10.1016/j.bpj.2010.12.3711
F. Palmieri, The mitochondrial transporter family ( SLC25 ): physiological and pathological implications, (2004) 689–709. https://doi.org/10.1007/s00424-003-1099-7.
Drew D, Boudker O (2016) Shared molecular mechanisms of membrane transporters. Annu Rev Biochem 85:543–572. https://doi.org/10.1146/annurev-biochem-060815-014520
Jardetzky O (1966) Simple allosteric model for membrane pumps. Nature 211:969–970. https://doi.org/10.1038/211969a0
Pebay-Peyroula E, Dahout-Gonzalez C, Kahn R, Trézéguet V, Lauquin GJ-M, Brandolin G (2003) Structure of mitochondrial ADP/ATP carrier in complex with carboxyatractyloside. Nature 426:39–44. https://doi.org/10.1038/nature02056
Nury H, Dahout-Gonzalez C, Trézéguet V, Lauquin G, Brandolin G, Pebay-Peyroula E (2005) Structural basis for lipid-mediated interactions between mitochondrial ADP/ATP carrier monomers. FEBS Lett 579:6031–6036. https://doi.org/10.1016/j.febslet.2005.09.061
Ruprecht JJ, Hellawell AM, Harding M, Crichton PG, McCoy AJ, Kunji ERSS (2014) Structures of yeast mitochondrial ADP/ATP carriers support a domain-based alternating-access transport mechanism. Proc Natl Acad Sci USA 111:E426–E434. https://doi.org/10.1073/pnas.1320692111
Ruprecht JJ, King MS, Zögg T, Aleksandrova AA, Pardon E, Crichton PG, Steyaert J, Kunji ERS (2019) The molecular mechanism of transport by the mitochondrial ADP/ATP carrier. Cell 176:435-447.e15. https://doi.org/10.1016/j.cell.2018.11.025
Ruprecht JJ, Kunji ERS (2020) The SLC25 mitochondrial carrier family: structure and mechanism. Trends Biochem Sci 45:244–258. https://doi.org/10.1016/j.tibs.2019.11.001
Palmieri F, Monné M (1863) Discoveries, metabolic roles and diseases of mitochondrial carriers: a review. Biochim Biophys Acta Mol Cell Res 2016:2362–2378. https://doi.org/10.1016/j.bbamcr.2016.03.007
Marobbio CMT, Vozza A, Harding M, Bisaccia F, Palmieri F, Walker JE (2002) Identification and reconstitution of the yeast mitochondrial transporter for thiamine pyrophosphate. EMBO J 21:5653–5661. https://doi.org/10.1093/emboj/cdf583
Best RB, Zhu X, Shim J, Lopes PEM, Mittal J, Feig M, MacKerell AD (2012) Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone ϕ, ψ and side-chain χ1 and χ2 dihedral angles. J Chem Theory Comput 8:3257–3273. https://doi.org/10.1021/ct300400x
Shelley JC, Cholleti A, Frye LL, Greenwood JR, Timlin MR, Uchimaya M (2007) Epik: A software program for pKa prediction and protonation state generation for drug-like molecules. J Comput Aided Mol Des 21:681–691. https://doi.org/10.1007/s10822-007-9133-z
Bright GR, Fisher GW, Rogowska J, Taylor DL (1987) Fluorescence ratio imaging microscopy: temporal and spatial measurements of cytoplasmic pH. J Cell Biol 104:1019–1033. https://doi.org/10.1083/jcb.104.4.1019
Santo-Domingo J, Demaurex N (2012) Perspectives on: SGP symposium on mitochondrial physiology and medicine: the renaissance of mitochondrial pH. J Gen Physiol 139:415–423. https://doi.org/10.1085/jgp.201110767
Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, Darian E, Guvench O, Lopes P, Vorobyov I, MacKerell AD (2010) CHARMM general force field: a force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J Comput Chem 31:671–690. https://doi.org/10.1002/jcc.21367
Vanommeslaeghe K, MacKerell AD (2012) Automation of the CHARMM general force field (CGenFF) I: bond perception and atom typing. J Chem Inf Model 52:3144–3154. https://doi.org/10.1021/ci300363c
Vanommeslaeghe K, Raman EP, MacKerell AD (2012) Automation of the CHARMM general force field (CGenFF) II: assignment of bonded parameters and partial atomic charges. J Chem Inf Model 52:3155–3168. https://doi.org/10.1021/ci3003649
Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graph 14:33–38. https://doi.org/10.1016/0263-7855(96)00018-5
Mayne CG, Saam J, Schulten K, Tajkhorshid E, Gumbart JC (2013) Rapid parameterization of small molecules using the force field toolkit. J Comput Chem 34:2757–2770. https://doi.org/10.1002/jcc.23422
Chauvet-Monges AM, Monti JP, Crevat A, Vincent EJ (1980) Étude de l’interaction de la thiamine diphosphate avec l’ion magnésium. Biochimie 61:1301–1308. https://doi.org/10.1016/S0300-9084(80)80289-6
Krämer R (1980) Influence of divalent cations on the reconstituted ADP, ATP exchange. Biochim Biophys Acta Bioenerg 592:615–620. https://doi.org/10.1016/0005-2728(80)90104-8
Gropp T, Brustovetsky N, Klingenberg M, Müller V, Fendler K, Bamberg E (1999) Kinetics of electrogenic transport by the ADP/ATP carrier. Biophys J 77:714–726. https://doi.org/10.1016/S0006-3495(99)76926-2
Gout E, Rébeillé F, Douce R, Bligny R (2014) Interplay of Mg2+, ADP, and ATP in the cytosol and mitochondria: unravelling the role of Mg2+ in cell respiration. Proc Natl Acad Sci USA 111:E4560–E4567. https://doi.org/10.1073/pnas.1406251111
Rostovtseva T, Colombini M (1997) VDAC channels mediate and gate the flow of ATP: implications for the regulation of mitochondrial function. Biophys J 72:1954–1962. https://doi.org/10.1016/S0006-3495(97)78841-6
Böckmann RA, De Groot BL, Kakorin S, Neumann E, Grubmüller H (2008) Kinetics, statistics, and energetics of lipid membrane electroporation studied by molecular dynamics simulations. Biophys J 95:1837–1850. https://doi.org/10.1529/BIOPHYSJ.108.129437
Vernier TP, Ziegler MJ (2007) Nanosecond field alignment of head group and water dipoles in electroporating phospholipid bilayers. J Phys Chem B 111:12993–12996. https://doi.org/10.1021/JP077148Q
Bolhuis PG (2006) Sampling kinetic protein folding pathways using all-atom models. Lect Notes Phys 703:393–433. https://doi.org/10.1007/3-540-35273-2_11
Villinger S, Giller K, Bayrhuber M, Lange A, Griesinger C, Becker S, Zweckstetter M (2014) Nucleotide interactions of the human voltage-dependent anion channel. J Biol Chem 289:13397–13406. https://doi.org/10.1074/jbc.M113.524173
Yehezkel G, Hadad N, Zaid H, Sivan S, Shoshan-Barmatz V (2006) Nucleotide-binding sites in the voltage-dependent anion channel: characterization and localization. J Biol Chem 281:5938–5946. https://doi.org/10.1074/jbc.M510104200
Nury H, Dahout-Gonzalez C, Trézéguet V, Lauquin GJM, Brandolin G, Pebay-Peyroula E (2006) Relations between structure and function of the mitochondrial ADP/ATP carrier. Annu Rev Biochem 75:713–741. https://doi.org/10.1146/annurev.biochem.75.103004.142747
Heimpel S, Basset G, Odoy S, Klingenberg M (2001) Expression of the mitochondrial ADP/ATP carrier in Escherichia coli. Renaturation, reconstitution, and the effect of mutations on 10 positive residues. J Biol Chem 276:11499–11506. https://doi.org/10.1074/jbc.M010586200
Liang J, Naveed H, Jimenez-Morales D, Adamian L, Lin M (1818) Computational studies of membrane proteins: models and predictions for biological understanding. Biochim Biophys Acta Biomembr 2012:927–941. https://doi.org/10.1016/j.bbamem.2011.09.026
Kunji ERS, Aleksandrova A, King MS, Majd H, Ashton VL, Cerson E, Springett R, Kibalchenko M, Tavoulari S, Crichton PG, Ruprecht JJ (1863) The transport mechanism of the mitochondrial ADP/ATP carrier. Biochim Biophys Acta Mol Cell Res 2016:2379–2393. https://doi.org/10.1016/j.bbamcr.2016.03.015
Brandolin G, Doussiere J, Gulik A, Gulik-Krzywicki T, Lauquin GJM, Vignais PV (1980) Kinetic, binding and ultrastructural properties of the beef heart adenine nucleotide carrier protein after incorporation into phospholipid vesicles. Biochim Biophys Acta Bioenerg 592:592–614. https://doi.org/10.1016/0005-2728(80)90103-6
Noskov SY, Rostovtseva TK, Chamberlin AC, Teijido O, Jiang W, Bezrukov SM (1858) Current state of theoretical and experimental studies of the voltage-dependent anion channel (VDAC). Biochim Biophys Acta Biomembr 2016:1778–1790. https://doi.org/10.1016/j.bbamem.2016.02.026
Robinson AJ, Overy C, Kunji ERS (2008) The mechanism of transport by mitochondrial carriers based on analysis of symmetry. Proc Natl Acad Sci 105:17766–17771. https://doi.org/10.1073/pnas.0809580105
Robinson AJ, Kunji ERS (2006) Mitochondrial carriers in the cytoplasmic state have a common substrate binding site. Proc Natl Acad Sci USA 103:2617–2622. https://doi.org/10.1073/pnas.0509994103
Mifsud J, Ravaud S, Krammer E-M, Chipot C, Kunji ERS, Pebay-Peyroula E, Dehez F (2013) The substrate specificity of the human ADP/ATP carrier AAC1. Mol Membr Biol 30:160–168. https://doi.org/10.3109/09687688.2012.745175
Wang Y, Tajkhorshid E (2008) Electrostatic funneling of substrate in mitochondrial inner membrane carriers. Proc Natl Acad Sci USA 105:9598–9603. https://doi.org/10.1073/pnas.0801786105
Bidon-Chanal A, Krammer E-M, Blot D, Pebay-Peyroula E, Chipot C, Ravaud S, Dehez F (2013) How do membrane transporters sense pH? The case of the mitochondrial ADP–ATP carrier. J Phys Chem Lett 4:3787–3791. https://doi.org/10.1021/jz401847d
Šponer J, Leszczynski J, Hobza P (2001) Electronic properties, hydrogen bonding, stacking, and cation binding of DNA and RNA bases. Biopolymers 61:3–31
Miniero DV, Cappello AR, Curcio R, Ludovico A, Daddabbo L, Stipani I, Robinson AJ, Kunji ERS, Palmieri F (1807) Functional and structural role of amino acid residues in the matrix α-helices, termini and cytosolic loops of the bovine mitochondrial oxoglutarate carrier. Biochim Biophys Acta Bioenerg 2011:302–310. https://doi.org/10.1016/j.bbabio.2010.12.005
Cappello AR, Miniero DV, Curcio R, Ludovico A, Daddabbo L, Stipani I, Robinson AJ, Kunji ERS, Palmieri F (2007) Functional and structural role of amino acid residues in the odd-numbered transmembrane α-helices of the bovine mitochondrial oxoglutarate carrier. J Mol Biol 369:400–412. https://doi.org/10.1016/j.jmb.2007.03.048
Klingenberg M (2005) Ligand-protein interaction in biomembrane carriers. The induced transition fit of transport catalysis, Biochemistry 44:8563–8570. https://doi.org/10.1021/bi050543r
Frisch GWTMJ, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JRJ, Scalmani G, Barone V, Mennucci B, Petersson GA, Nakatsuji H, Caricato M, Li X, Hratchian HP, Izmaylov AF, Bloino J, Zheng G, Sonnenberg JL, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Vreven T, Montgomery JJA, Peralta JE, Ogliaro F, Bearpark M, Heyd JJ, Brothers E, Kudin KN, Staroverov VN, Kobayashi R, Normand J, Raghavachari K, Rendell A, Burant JC, Iyengar SS, Tomasi J, Cossi M, Rega N, Millam JM, Klene M, Knox JE, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin VJ, AJ, Cioslowski DJ, Gaussian 09, Revision A.1, Vol. Gaussian, Inc., Wallingford CT (2009)
Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K (2005) Scalable molecular dynamics with NAMD. J Comput Chem 26:1781–1802. https://doi.org/10.1002/jcc.20289
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410. https://doi.org/10.1016/S0022-2836(05)80360-2
Söding J, Biegert A, Lupas AN (2005) The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. https://doi.org/10.1093/nar/gki408
Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. https://doi.org/10.1038/msb.2011.75
Tusnady GE, Simon I (2001) The HMMTOP transmembrane topology prediction server. Bioinformatics 17:849–850. https://doi.org/10.1093/bioinformatics/17.9.849
Hofmann K, Stoffel W (1993) TMbase-A database of membrane spanning proteins segments. Biol Chem 374:166
Šali A, Blundell TL (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234:779–815
Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26:283–291. https://doi.org/10.1107/S0021889892009944
MacKerell AD, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FT, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiórkiewicz-Kuczera J, Yin D, Karplus M (1998) All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B 102:3586–3616. https://doi.org/10.1021/jp973084f
Lomize MA, Lomize AL, Pogozheva ID, Mosberg HI (2006) OPM: Orientations of proteins in membranes database. Bioinformatics 22:623–625. https://doi.org/10.1093/bioinformatics/btk023
Lee J, Cheng X, Swails JM, Yeom MS, Eastman PK, Lemkul JA, Wei S, Buckner J, Jeong JC, Qi Y, Jo S, Pande VS, Case DA, Brooks CL, MacKerell AD, Klauda JB, Im W (2015) CHARMM-GUI input generator for NAMD, GROMACS, AMBER, OpenMM, and CHARMM/OpenMM simulations using the CHARMM36 additive force field. J Chem Theory Comput. https://doi.org/10.1021/acs.jctc.5b00935
Tomasello MF, Guarino F, Reina S, Messina A, De Pinto V (2013) The voltage-dependent anion selective channel 1 (VDAC1) topography in the mitochondrial outer membrane as detected in intact cell. PLoS ONE 8:e81522. https://doi.org/10.1371/journal.pone.0081522
Watson HC, Walker NP, Shaw PJ, Bryant TN, Wendell PL, Fothergill LA, Perkins RE, Conroy SC, Dobson MJ, Tuite MF (1982) Sequence and structure of yeast phosphoglycerate kinase. EMBO J 1:1635–1640. https://doi.org/10.1002/j.1460-2075.1982.tb01366.x
Aksimentiev A, Schulten K (2005) Imaging alpha-hemolysin with molecular dynamics: ionic conductance, osmotic permeability, and the electrostatic potential map. Biophys J 88:3745–3761. https://doi.org/10.1529/biophysj.104.058727
Tsoulos IG, Stavrakoudis A (2011) Eucb: A C++ program for molecular dynamics trajectory analysis. Comput Phys Commun 182:834–841. https://doi.org/10.1016/j.cpc.2010.11.032
Dehez F, Pebay-Peyroula E, Chipot C (2008) Binding of ADP in the mitochondrial ADP/ATP carrier is driven by an electrostatic funnel. J Am Chem Soc 130:12725–12733. https://doi.org/10.1021/ja8033087
Krammer E-M, Ravaud S, Dehez F, Frelet-Barrand A, Pebay-Peyroula E, Chipot C (2009) High-chloride concentrations abolish the binding of adenine nucleotides in the mitochondrial ADP/ATP carrier family. Biophys J 97:L25–L27. https://doi.org/10.1016/j.bpj.2009.08.047
Kang J, Samuels DC (2008) The evidence that the DNC (SLC25A19 ) is not the mitochondrial deoxyribonucleotide carrier. 8:103–108. https://doi.org/10.1016/j.mito.2008.01.001
Smart OS, Neduvelil JG, Wang X, Wallace BA, Sansom MSP (1996) HOLE: a program for the analysis of the pore dimensions of ion channel structural models. J Mol Gr 14:354–360. https://doi.org/10.1016/S0263-7855(97)00009-X
Acknowledgements
M.P. is a senior research associate and E.M.K. is a postdoctoral researcher of the Fonds de la Recherche Scientifique de Belgique (F.R.S.-F.R.N.S.), Belgium. Computational resources were provided by the Consortium des Équipements de Calcul Intensif (CÉCI) and the F.R.S.-F.N.R.S. under convention 2.5020.11, together with the supercomputing facilities of the Université catholique de Louvain (CISM/UCL), the Université de Liège (ULg) and ULB.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Van Liefferinge, F., Krammer, EM., Waeytens, J. et al. Molecular mechanism of thiamine pyrophosphate import into mitochondria: a molecular simulation study. J Comput Aided Mol Des 35, 987–1007 (2021). https://doi.org/10.1007/s10822-021-00414-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10822-021-00414-5