Abstract
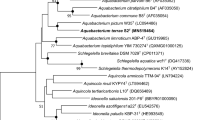
A novel Gram-stain-negative, rod-shaped, strictly aerobic, non-motile bacterium, designated strain cd-1T, was isolated from a farmland soil applied with amino acid fertilizer in Zhengzhou, Henan province, China. The optimum growth of strain cd-1T occurred at 30 °C, pH 7.0 in Luria–Bertani (LB) broth without NaCl supplement. Phylogenetic analysis based on 16S rRNA gene sequences indicated that cd-1T is member of the genus Aquamicrobium, and formed a separate branch with Aquamicrobium aerolatum DSM 21857T (96.5%) and Aquamicrobium soli KCTC 52165T (95.7%). The draft genome sequencing revealed a DNA G + C content of 59.2 mol% and Q-10 was the predominant respiratory quinone. The major cellular fatty acids were identified as C18:1 ω7c (35.8%), C19:0 cyclo ω8c (32.1%), and C18:1 ω7c 11-methyl (5.2%). The polar lipids consisted of phosphatidylethanolamine, diphosphatidylglycerol, phosphatidylglycerol, phosphatidylcholine and phosphatidylmonomethylethanolamine. Average nucleotide identity (ANI) and the digital DNA-DNA hybridizations (dDDH) for draft genomes between strain cd-1T and KCTC 52165T were 71.0% and 19.9%, respectively, the values for strain cd-1T and DSM 21857T were 73.4% and 20.6%. Based on the physiological and biochemical characteristics, phylogenetic and chemotaxonomic analysis, strain cd-1T is considered to represent a novel species of the genus Aquamicrobium, for which the name Aquamicrobium zhengzhouense sp. nov. is proposed. The type strain is cd-1T (= KCTC 82182T = CCTCC M 2018904T).

Similar content being viewed by others
References
Bambauer A, Rainey FA, Stackebrandt E, Winter J (1998) Characterization of Aquamicrobium defluvii gen. nov. sp. nov., a thiophene-2-carboxylate-metabolizing bacterium from activated sludge. Arch Microbiol 169:293–302
Kampfer P, Martin E, Lodders N, Jackel U (2009) Transfer of Defluvibacter lusatiensis to the genus Aquamicrobium as Aquamicrobium lusatiense comb. nov. and description of Aquamicrobium aerolatum sp. nov. Int J Syst Evol Microbiol 59:2468–2470
Jin HM, Kim JM, Jeon CO (2013) Aquamicrobium aestuarii sp. nov., a marine bacterium isolated from a tidal flat. Int J Syst Evol Microbiol 63:4012–4017
Lipski A, Kämpfer P (2012) Aquamicrobium ahrensii sp. nov. and Aquamicrobium segne sp. nov., isolated from experimental biofilters. Int J Syst Evol Microbiol 62:2511–2516
Xu C, Zhang L, Huang J, Chen K, Li S, Jiang J (2017) Aquamicrobium soli sp. nov., a bacterium isolated from a chlorobenzoate-contaminated soil. Anton Leeuw Int J G 110:305–312
Wu Z, Wang F, Gu C, Zhang Y, Yang Z, Wu X, Jiang X (2014) Aquamicrobium terrae sp. nov., isolated from the polluted soil near a chemical factory. Anton Leeuw Int J G 105:1131–1137
Marchesi JR, Sato T, Weightman AJ, Martin TA, Fry JC, Hiom SJ, Wade WG (1998) Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl Environ Microbiol 64:795–799
Kim OS, Cho Y, Lee K, Yoon SH, Kim M, Na H, Park SC, Jeon Y, Lee JH, Yi H (2012) Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol 62:716–721
Thompson JD, Gibson TJ, Frédéric P, Franois J, Higgins DG (1997) The clustal_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA 6.0: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Saitou N, Nei M (1987) The neighbor-joining methods: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Fitch Walter M (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Syst Biol 20:406–416
Felsenstein J (2013) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Stackebrandt E, Goebel BM (1994) Taxonomic note: a place for DNA-DNA reassociation and 16S rRNA sequence analysis in the present species definition in bacteriology. Int J Syst Evol Microbiol 44:846–849
Bernardet J, Nakagawa Y, Holmes B (2002) Proposed minimal standards for describing new taxa of the family Flavobacteriaceae and emended description of the family. Int J Syst Evol Microbiol 52:1049–1070
Collins MD, Pirouz T, Goodfellow M, Minnikin DE (1977) Distribution of Menaquinones in Actinomycetes and Corynebacteria. J Gen Microbiol 100:221–230
Li R, Zhu H, Ruan J, Qian W, Fang X, Shi Z, Li Y, Li S, Shan G, Kristiansen K (2010) De novo assembly of human genomes with massively parallel short read sequencing. Genome Res 20:265–272
Meier-Kolthoff JP, Auch AF, Klenk H, Göker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform 14:60–60
Chun J, Oren A, Ventosa A, Christensen H, Arahal DR, Costa MSD, Rooney AP, Yi H, Xu X, De Meyer SE (2018) Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol 68:461–466
Funding
Key research and development projects in Anhui Province Grant No. (202004f06020049). Major Science and Technology Projects in Anhui Province Grant No. (18030701214). Natural Science Foundation of the Anhui Higher Education Institutions of China Grant Nos. (KJ2020A0050, KJ2018A0530). Anhui Province University Student Innovation and Entrepreneurship Training Program Project Grant No. (S201910879040). Stable Talent Project of Anhui Science and Technology University.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Research Involving Human Participants and/or Animals
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, X., Zhou, CF., Cheng, JR. et al. Aquamicrobium zhengzhouense sp. nov., a Bacterium Isolated from Farmland Soil Applied with Amino Acid Fertilizer. Curr Microbiol 78, 3798–3803 (2021). https://doi.org/10.1007/s00284-021-02600-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-021-02600-y