Abstract
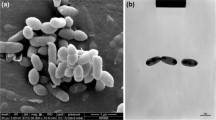
A novel actinobacterial strain, Gram-positive, anaerobic, non-motile, and rod-shaped, designated KGMB02528T, was isolated from healthy human feces. Cells of strain KGMB02528T grew optimally at pH 7.0 and 37 °C and in the presence of 0% (w/v) NaCl. Based on 16S rRNA gene sequence similarity, strain KGMB04489T belonged to the genus Collinsella and was most closely related to Collinsella aerofaciens DSM 17552T (95.8%). The DNA G + C content was 58.0 mol%. The major cellular fatty acids (> 10%) were C16:0 DMA, C16:0 ALDE, C14:0 DMA, and C12:0. The predominant end product of fermentation was acetic acid. The cell wall peptidoglycan of strain KGMB02528T contained alanine, glutamic acid, and lysine, while diaminopimelic acid was not detected. The polar lipids were composed of two unidentified phospholipids and unidentified nine glycolipids. Based on the phenotypic, chemotaxonomic, and phylogenetic properties, strain KGMB02528T represents a novel species of the genus Collinsella, for which the name Collinsella acetigenes sp. nov. is proposed. The type strain is Collinsella acetigenes KGMB02528T (= KCTC 15847T = CCUG 73987T). The description of the genus Collinsella is emended to accommodate the new species.
The GenBank/EMBL/DDBJ accession number for the 16S rRNA gene sequence of Collinsella acetigenes KGMB02528T is MT117838. The whole-genome shotgun BioProject number is PRJNA623694 with the accession number JABBCP000000000.

Similar content being viewed by others
Data Availability
We agree to the transparency of the material. Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study, or all data generated or analyzed during this study are included in this published article [and its supplementary information files].
Abbreviations
- ALDE:
-
Aldehyde
- ANI:
-
Average nucleotide identity
- CCUG:
-
Culture Collection University of Gothenburg
- DAP:
-
Diaminopimelic acid
- dDDH:
-
digital DNA–DNA hybridization
- DMA:
-
Dimethyl acetate
- GL:
-
Glycolipid
- HPLC:
-
High-performance liquid chromatography
- KCTC:
-
Korean Collection for Type Cultures
- PL:
-
Phospholipid
- TLC:
-
Thin-layer chromatography
References
Stackebrandt E, Rainey FA, Ward-Rainey NL (1997) Proposal for a new hierarchic classification system, actinobacteria classis nov. Int J Syst Bacteriol 47:479–491
Kageyama A, Benno Y, Nakase T (1999) Phylogenetic and phenotypic evidence for the transfer of Eubacterium aerofaciens to the genus Collinsella as Collinsella aerofaciens gen. nov., comb. nov. Int J Syst Bacteriol 49:557–565
Euzéby JP (2018) LPSN—List of prokaryotic names with standing in nomenclature (bacterio.net), 20 years on. Int J Syst Bacteriol 68:1825–1829
Kageyama A, Benno Y (2000) Emendation of genus Collinsella and proposal of Collinsella stercoris sp. nov. and Collinsella intestinalis sp. nov. Int J Syst Evol Microbiol 50:1767–1774
Padmanabhan R, Dubourg G, Lagier JC, Nguyen TT, Couderc C, Rossi-Tamisier M, Caputo A, Raoult D, Fournier PE (2014) Non-contiguous finished genome sequence and description of Collinsella massiliensis sp. nov. Stand Genomic Sci 9:1144–1158
Diop A, Diop K, Tomei E, Armstrong N, Bretelle F, Raoult D, Fenollar F, Fournier PE (2019) Collinsella vaginalis sp. nov. strain Marseille-P2666T, a new member of the Collinsella genus isolated from the genital tract of a patient suffering from bacterial vaginosis. Int J Syst Evol Microbiol 69:949–956
Yoon SH, Ha SM, Kwon S, Lim J, Kim Y, Seo H, Chun J (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67:1613–1617
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41:95–98
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Fitch WM (1971) Toward defining the course of evolution: minimum change for a specific tree topology. Syst Zool 20:406–416
Felsenstein J (1981) Evolutionary trees from DNA sequences: a maximum likelihood approach. J Mol Evol 17:368–376
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477
Lee I, Chalita M, Ha SM, Na SI, Yoon SH, Chun J (2017) ContEst16S: an algorithm that identifies contaminated prokaryotic genomes using 16S RNA gene sequences. Int J Syst Evol Microbiol 67:2053–2057
Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW (2015) CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res 25:1043–1055
Yoon SH, Ha SM, Lim J, Kwon S, Chun J (2017) A large-scale evaluation of algorithms to calculate average nucleotide identity. Antonie Van Leeuwenhoek 110:1281–1286
Meier-Kolthoff JP, Auch AF, Klenk HP, Göker M (2013) Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinform 14:60
Sasser M (1990) Identification of bacteria by gas chromatography of cellular fatty acids, MIDI technical note 101. MIDI Inc, Newark
Komagata K, Suzuki K-I (1987) Lipid and cell-wall analysis on bacterial systematics. In: Colwell RR, Grigorova R (eds) Methods in microbiology, vol 19. Academic Press, London, pp 177–182
Staneck JL, Roberts GD (1974) Simplified approach to identification of aerobic actinomycetes by thin-layer chromatography. Appl Microbiol 28:226–231
Schleifer KH, Kandler O (1972) Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol Rev 36:407–477
Zhang X, Rimpiläinen M, Simelyte E, Toivanen P (2000) What determines arthritogenicity of bacterial cell wall? A study on Eubacterium cell wall-induced arthritis. Rheumatology 39:274–282
Goris J, Konstantinidis KT, Klappenbach JA, Coenye T, Vandamme P, Tiedje JM (2007) DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol 57:81–91
Richter M, Rosselló-Móra R (2009) Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci USA 106:19126–19131
Nagai F, Watanabe Y, Morotomi M (2010) Slackia piriformis sp. nov. and Collinsella tanakaei sp. nov., new members of the family Coriobacteriaceae, isolated from human faeces. Int J Syst Evol Microbiol 60:2639–2646
Acknowledgements
We thank Professor Bernhard Schink for his advice in naming the novel strain.
Funding
This work was supported by the Bio & Medical Technology Development program (Project No. NRF-2016H3A9F3947962) of the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT (MSIT) of the Republic of Korea and a grant from the Korea Research Institute of Bioscience & Biotechnology (KRIBB) Research initiative program.
Author information
Authors and Affiliations
Contributions
All authors participating in this study were supported by NRF-2016H3A9F3947962 [Supervision: JSL]. Sampling by the IRB: DHL, HY. Gut microbial isolation and taxonomy analysis were performed by JSK, MKE, KCL, MKS, HSK, SHP, JHL, SWK, JEP, BSO, SWR, SYY, and SHC. Gut microbial whole-genome analysis was performed by BYK, JHL. The first draft of the manuscript was written by KIH and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript. Conceptualization: JSL. Formal analysis and investigation: KIH and MKE. Writing—original draft preparation: KIH. Writing—revised draft preparation and experiment: JSK. Review and editing: JSL. Funding acquisition: JSL. Resources: JSK, KCL, MKS, HSK, SHP, JHL, SWK, JEP, BSO, SWR, SYY, SHC, DHL, HY, BYK, JHL. Supervision: JSL.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical Approval
The study was approved by the Institutional Review Board (IRB) of Korea Research Institute of Bioscience & Biotechnology (Approval number: P01-201702-31-007).
Consent to Participate
All authors agree to participate.
Consent for Publication
All authors agree to the publication.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Han, KI., Kim, JS., Eom, M.K. et al. Collinsella acetigenes sp. nov., an Anaerobic Actinobacterium Isolated from Human Feces, and Emended Description of the Genus Collinsella and Collinsella aerofaciens. Curr Microbiol 78, 3667–3673 (2021). https://doi.org/10.1007/s00284-021-02625-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00284-021-02625-3