Abstract
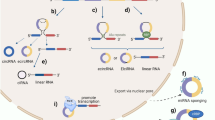
Circular RNAs (circRNAs) are a class of endogenously expressed regulatory RNAs with a single-stranded circular structure. They are generated by back splicing and their expression can be tightly regulated by RNA binding proteins. Cytoplasmic circRNAs can function as molecular sponges that inhibit microRNA–target interactions and protein function or as templates for the efficient generation of peptides via rolling circle amplification. They can also act as molecular scaffolds that enhance the reaction kinetics of enzyme–substrate interactions. In the nucleus, circRNAs might facilitate chromatin modifications and promote gene expression. CircRNAs are resistant to degradation and can be packaged in extracellular vesicles and transported in the circulation. Initial studies suggest that circRNAs have roles in kidney disease and associated cardiovascular complications. They have been implicated in hypertensive nephropathy, diabetic kidney disease, glomerular disease, acute kidney injury and kidney allograft rejection, as well as in microvascular and macrovascular complications of chronic kidney disease, including atherosclerotic vascular disease. In addition, several circRNAs have been reported to have oncogenic or tumour suppressor roles or to regulate drug resistance in kidney cancer. The available data suggest that circRNAs could be promising diagnostic and/or prognostic biomarkers and potential therapeutic targets for kidney disease, cardiovascular disease and kidney cancer.
Key points
-
CircRNAs are regulatory RNA molecules with a closed circular structure that are generated by back splicing of precursor mRNAs.
-
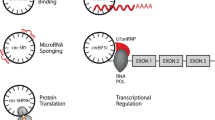
Functions of cytoplasmic circRNAs include sponging of microRNAs and proteins, scaffolding of enzyme–substrate interactions and acting as templates for protein translation.
-
Nuclear-enriched circRNAs can also act as molecular sponges and promote gene expression by interacting with chromatin remodelling complexes and increasing RNA polymerase II activity.
-
CircRNAs have been implicated in the pathogenesis of kidney diseases, cardiovascular complications of chronic kidney disease and kidney cancer, and are promising potential therapeutic targets.
-
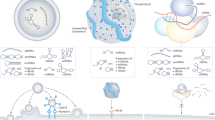
CircRNAs are promising biomarkers of disease owing to their high stability and packaging in extracellular vesicles.
-
Potential circRNA-based therapeutic approaches include modulation of native circRNAs and the application of artificial circRNAs with designer molecular functions.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Change history
31 August 2021
The formatting of Anton Jan van Zonneveld’s name in the xml was incorrect, resulting in Jan being wrongly classed as part of his surname. This has now been corrected online.
References
Salzman, J., Gawad, C., Wang, P. L., Lacayo, N. & Brown, P. O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 7, e30733 (2012).
Jeck, W. R. et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19, 141–157 (2013).
Memczak, S. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495, 333–338 (2013).
Hansen, T. B. et al. Natural RNA circles function as efficient microRNA sponges. Nature 495,384–388 (2013).
Sanger, H. L., Klotz, G., Riesner, D., Gross, H. J. & Kleinschmidt, A. K. Viroids are single-stranded covalently closed circular RNA molecules existing as highly base-paired rod-like structures. Proc. Natl Acad. Sci. USA 73, 3852–3856 (1976).
Berget, S. M., Moore, C. & Sharp, P. A. Spliced segments at the 5′ terminus of adenovirus 2 late mRNA. Proc. Natl Acad. Sci. USA 74, 3171–3175 (1977).
Nigro, J. M. et al. Scrambled exons. Cell 64, 607–613 (1991).
Kos, A., Dijkema, R., Arnberg, A. C., van der Meide, P. H. & Schellekens, H. The hepatitis delta (delta) virus possesses a circular RNA. Nature 323, 558–560 (1986).
Cocquerelle, C., Mascrez, B., Hetuin, D. & Bailleul, B. Mis-splicing yields circular RNA molecules. FASEB J. 7, 155–160 (1993).
Bailleul, B. During in vivo maturation of eukaryotic nuclear mRNA, splicing yields excised exon circles. Nucleic Acids Res. 24, 1015–1019 (1996).
Zhang, X. O. et al. Complementary sequence-mediated exon circularization. Cell 159, 134–147 (2014).
Ashwal-Fluss, R. et al. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell 56, 55–66 (2014).
Conn, S. J. et al. The RNA binding protein quaking regulates formation of circRNAs. Cell 160,1125–1134 (2015).
Salzman, J., Chen, R. E., Olsen, M. N., Wang, P. L. & Brown, P. O. Cell-type specific features of circular RNA expression. PLoS Genet. 9, e1003777 (2013).
Delaleau, M. & Borden, K. L. Multiple export mechanisms for mRNAs. Cells 4, 452–473 (2015).
Huang, C., Liang, D., Tatomer, D. C. & Wilusz, J. E. A length-dependent evolutionarily conserved pathway controls nuclear export of circular RNAs. Genes Dev. 32, 639–644 (2018).
Hansen, T. B. et al. miRNA-dependent gene silencing involving Ago2-mediated cleavage of a circular antisense RNA. EMBO J. 30, 4414–4422 (2011).
Park, O. H. et al. Endoribonucleolytic cleavage of m6A-containing RNAs by RNase P/MRP complex. Mol. Cell 74, 494–507.e8 (2019).
Lee, Y., Choe, J., Park, O. H. & Kim, Y. K. Molecular mechanisms driving mRNA degradation by m(6)A modification. Trends Genet. 36, 177–188 (2020).
Liu, C. X. et al. Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell 177, 865–80.e21 (2019).
Fischer, J. W., Busa, V. F., Shao, Y. & Leung, A. K. L. Structure-mediated RNA decay by UPF1 and G3BP1. Mol. Cell 78, 70–84.e6 (2020).
Li, Y. et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 25, 981–984 (2015).
Preusser, C. et al. Selective release of circRNAs in platelet-derived extracellular vesicles.J. Extracell. Vesicles 7, 1424473 (2018).
Lasda, E. & Parker, R. Circular RNAs co-precipitate with extracellular vesicles: a possible mechanism for circRNA clearance. PLoS One 11, e0148407 (2016).
Raposo, G. & Stoorvogel, W. Extracellular vesicles: exosomes, microvesicles, and friends. J. Cell Biol. 200, 373–383 (2013).
Memczak, S., Papavasileiou, P., Peters, O. & Rajewsky, N. Identification and characterization of circular RNAs as a new class of putative biomarkers in human blood. PLoS One 10, e0141214 (2015).
van Balkom, B. W., Pisitkun, T., Verhaar, M. C. & Knepper, M. A. Exosomes and the kidney: prospects for diagnosis and therapy of renal diseases. Kidney Int. 80, 1138–1145 (2011).
Kristensen, L. S. et al. The biogenesis, biology and characterization of circular RNAs. Nat. Rev. Genet. 20, 675–691 (2019).
Piwecka, M. et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 357, eaam8526 (2017).
Kleaveland, B., Shi, C. Y., Stefano, J. & Bartel, D. P. A network of noncoding regulatory RNAs Acts in the mammalian brain. Cell 174, 350–62.e17 (2018).
Dudekula, D. B. et al. CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 13, 34–42 (2016).
Abdelmohsen, K. et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 14, 361–369 (2017).
Holdt, L. M. et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat. Commun. 7, 12429 (2016).
Du, W. W. et al. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 44, 2846–2858 (2016).
Pamudurti, N. R. et al. Translation of CircRNAs. Mol. Cell 66, 9–21.e7 (2017).
Legnini, I. et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell 66, 22–37.e9 (2017).
Zhang, M. et al. A peptide encoded by circular form of LINC-PINT suppresses oncogenic transcriptional elongation in glioblastoma. Nat. Commun. 9, 4475 (2018).
Zhang, M. et al. A novel protein encoded by the circular form of the SHPRH gene suppresses glioma tumorigenesis. Oncogene 37, 1805–1814 (2018).
Yang, Y. et al. Novel role of FBXW7 circular RNA in repressing glioma tumorigenesis. J. Natl Cancer Inst. 110, 304–315 (2018).
Chen, X. et al. circRNADb: a comprehensive database for human circular RNAs with protein-coding annotations. Sci. Rep. 6, 34985 (2016).
Xia, P. et al. A circular RNA protects dormant hematopoietic stem cells from DNA sensor cGAS-mediated exhaustion. Immunity 48, 688–701.e7 (2018).
Chen, N. et al. A novel FLI1 exonic circular RNA promotes metastasis in breast cancer by coordinately regulating TET1 and DNMT1. Genome Biol. 19, 218 (2018).
Zhang, Y. et al. Circular intronic long noncoding RNAs. Mol. Cell 51, 792–806 (2013).
Li, Z. et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 22, 256–264 (2015).
Lu, C. et al. CircNr1h4 regulates the pathological process of renal injury in salt-sensitive hypertensive mice by targeting miR-155-5p. J. Cell Mol. Med. 24, 1700–1712 (2020).
Cheng, X. & Joe, B. Circular RNAs in rat models of cardiovascular and renal diseases. Physiol. Genomics 49, 484–490 (2017).
Hu, W., Han, Q., Zhao, L. & Wang, L. Circular RNA circRNA_15698 aggravates the extracellular matrix of diabetic nephropathy mesangial cells via miR-185/TGF-beta1. J. Cell Physiol. 234, 1469–1476 (2019).
Wen, S., Li, S., Li, L. & Fan, Q. circACTR2: a novel mechanism regulating high glucose-induced fibrosis in renal tubular cells via pyroptosis. Biol. Pharm. Bull. 43, 558–564 (2020).
Guo, G. et al. Hsa_circ_0000479 as a novel diagnostic biomarker of systemic lupus erythematosus. Front. Immunol. 10, 2281 (2019).
Luan, J. et al. circHLA-C plays an important role in lupus nephritis by sponging miR-150. Mol. Ther. Nucleic Acids 10, 245–253 (2018).
Jin, X. et al. Comprehensive expression profiles and bioinformatics analysis reveal special circular RNA expression and potential predictability in the peripheral blood of humans with idiopathic membranous nephropathy. Mol. Med. Rep. 20, 4125–4139 (2019).
Cao, Y. et al. Transcriptome sequencing of circular RNA reveals a novel circular RNA-has_circ_0114427 in the regulation of inflammation in acute kidney injury. Clin. Sci. 134, 139–154 (2020).
Huang, T. et al. Circular RNA YAP1 acts as the sponge of microRNA-21-5p to secure HK-2 cells from ischaemia/reperfusion-induced injury. J. Cell Mol. Med. 24, 4707–4715 (2020).
Kolling, M. et al. The circular RNA ciRs-126 predicts survival in critically Ill patients with acute kidney injury. Kidney Int. Rep. 3, 1144–1152 (2018).
Dou, Y. Q. et al. Smooth muscle SIRT1 reprograms endothelial cells to suppress angiogenesis after ischemia. Theranostics 10, 1197–1212 (2020).
Kolling, M. et al. Circular RNAs in urine of kidney transplant patients with acute T cell-mediated allograft rejection. Clin. Chem. 65, 1287–1294 (2019).
Altesha, M. A., Ni, T., Khan, A., Liu, K. & Zheng, X. Circular RNA in cardiovascular disease. J. Cell Physiol. 234, 5588–5600 (2019).
Aufiero, S., Reckman, Y. J., Pinto, Y. M. & Creemers, E. E. Circular RNAs open a new chapter in cardiovascular biology. Nat. Rev. Cardiol. 16, 503–514 (2019).
Tonelli, M. et al. Risk of coronary events in people with chronic kidney disease compared with those with diabetes: a population-level cohort study. Lancet. 380, 807–814 (2012).
McCullough, P. A. Why is chronic kidney disease the “spoiler” for cardiovascular outcomes? J. Am. Coll. Cardiol. 41, 725–728 (2003).
Bernelot Moens, S. J. et al. Arterial and cellular inflammation in patients with CKD. J. Am. Soc. Nephrol. 28, 1278–1285 (2017).
Malyszko, J. Mechanism of endothelial dysfunction in chronic kidney disease. Clin. Chim. Acta 411, 1412–1420 (2010).
Huang, H. S., Huang, X. Y., Yu, H. Z., Xue, Y. & Zhu, P. L. Circular RNA circ-RELL1 regulates inflammatory response by miR-6873-3p/MyD88/NF-kappaB axis in endothelial cells. Biochem. Biophys. Res. Commun. 525, 512–519 (2020).
Zhang, F. et al. Comprehensive analysis of circRNA expression pattern and circRNA-miRNA-mRNA network in the pathogenesis of atherosclerosis in rabbits. Aging 10, 2266–2283 (2018).
Burd, C. E. et al. Expression of linear and novel circular forms of an INK4/ARF-associated non-coding RNA correlates with atherosclerosis risk. PLoS Genet. 6, e1001233 (2010).
Goodman, W. G. et al. Coronary-artery calcification in young adults with end-stage renal disease who are undergoing dialysis. N. Engl. J. Med. 342, 1478–1483 (2000).
Cozzolino, M. et al. Cardiovascular disease in dialysis patients. Nephrol. Dial. Transplant. 33(Suppl_3), iii28–iii34 (2018).
Ryu, J. et al. Characterization of circular RNAs in vascular smooth muscle cells with vascular calcification. Mol. Ther. Nucleic Acids 19, 31–41 (2020).
Wang, Y. et al. Melatonin ameliorates aortic valve calcification via the regulation of circular RNA CircRIC3/miR-204-5p/DPP4 signaling in valvular interstitial cells. J. Pineal. Res. 69, e12666 (2020).
Koch, B. C. et al. Impairment of endogenous melatonin rhythm is related to the degree of chronic kidney disease (CREAM study). Nephrol. Dial. Transpl. 25, 513–519 (2010).
Karasek, M., Szuflet, A., Chrzanowski, W., Zylinska, K. & Swietoslawski, J. Decreased melatonin nocturnal concentrations in hemodialyzed patients. Neuro Endocrinol. Lett. 26, 653–656 (2005).
Armulik, A., Abramsson, A. & Betsholtz, C. Endothelial/pericyte interactions. Circ. Res. 97, 512–523 (2005).
Jiang, Q. et al. Circular RNA-ZNF532 regulates diabetes-induced retinal pericyte degeneration and vascular dysfunction. J. Clin. Invest. 130, 3833–3847 (2020).
Goligorsky, M. S. Pathogenesis of endothelial cell dysfunction in chronic kidney disease: a retrospective and what the future may hold. Kidney Res. Clin. Pract. 34, 76–82 (2015).
Yang, L. et al. Engagement of circular RNA HECW2 in the nonautophagic role of ATG5 implicated in the endothelial-mesenchymal transition. Autophagy 14, 404–418 (2018).
Kimura, T. et al. Autophagy protects the proximal tubule from degeneration and acute ischemic injury. J. Am. Soc. Nephrol. 22, 902–913 (2011).
Yousefi, F. & Soltani, B. M. Circular RNAs as potential theranostics in the cardiac fibrosis. Heart Fail Rev. 13, 407–418 (2020).
Wang, Y. & Liu, B. Circular RNA in diseased heart. Cells. 9, 1240 (2020).
Amann, K., Rychlik, I., Miltenberger-Milteny, G. & Ritz, E. Left ventricular hypertrophy in renal failure. Kidney Int. Suppl. 68, S78–S85 (1998).
McMullen, J. R. & Ooi, J. Y. Y. The interplay of protein coding and non-coding RNAs (circRNAs, lncRNAs) during cardiac differentiation. EBioMedicine 25, 9–10 (2017).
Tan, W. L. et al. A landscape of circular RNA expression in the human heart. Cardiovasc. Res. 113, 298–309 (2017).
Wang, K. et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur. Heart J. 37, 2602–2611 (2016).
Lim, T. B. et al. Targeting the highly abundant circular RNA circSlc8a1 in cardiomyocytes attenuates pressure overload induced hypertrophy. Cardiovasc. Res. 115, 1998–2007 (2019).
Zhao, X., Cai, Y. & Xu, J. Circular RNAs: biogenesis, mechanism, and function in human cancers. Int. J. Mol. Sci. 20, 3926 (2019).
Yu, T. et al. CircRNAs in cancer metabolism: a review. J. Hematol. Oncol. 12, 90 (2019).
Kalluri, R. & LeBleu, V. S. The biology, function, and biomedical applications of exosomes. Science 367, eaau6977 (2020).
Shi, X. et al. circRNAs and exosomes: a mysterious frontier for human cancer. Mol. Ther. Nucleic Acids 19, 384–392 (2020).
Chen, T., Shao, S., Li, W., Liu, Y. & Cao, Y. The circular RNA hsa-circ-0072309 plays anti-tumour roles by sponging miR-100 through the deactivation of PI3K/AKT and mTOR pathways in the renal carcinoma cell lines. Artif. Cell Nanomed. Biotechnol. 47, 3638–3648 (2019).
Zhou, B. et al. CircPCNXL2 sponges miR-153 to promote the proliferation and invasion of renal cancer cells through upregulating ZEB2. Cell Cycle 17, 2644–2654 (2018).
Zhang, D. et al. Down-regulation of circular RNA_000926 attenuates renal cell carcinoma progression through miRNA-411-dependent CDH2 inhibition. Am. J. Pathol. 189, 2469–2486 (2019).
Chen, Z. et al. Circular RNA hsa_circ_001895 serves as a sponge of microRNA-296-5p to promote clear cell renal cell carcinoma progression by regulating SOX12. Cancer Sci. 111, 713–726 (2020).
Xue, D. et al. Circ-AKT3 inhibits clear cell renal cell carcinoma metastasis via altering miR-296-3p/E-cadherin signals. Mol. Cancer 18, 151 (2019).
Han, Z. et al. ERbeta-mediated alteration of circATP2B1 and miR-204-3p signaling promotes invasion of clear cell renal cell carcinoma. Cancer Res. 78, 2550–2563 (2018).
Chen, Q. et al. CircRNA cRAPGEF5 inhibits the growth and metastasis of renal cell carcinoma via the miR-27a-3p/TXNIP pathway. Cancer Lett. 469, 68–77 (2020).
Franz, A. et al. Circular RNAs in clear cell renal cell carcinoma: their microarray-based identification, analytical validation, and potential use in a clinico-genomic model to improve prognostic accuracy. Cancers 11, 1473 (2019).
Lin, L. & Cai, J. Circular RNA circ-EGLN3 promotes renal cell carcinoma proliferation and aggressiveness via miR-1299-mediated IRF7 activation. J. Cell Biochem. 121, 4377–4385 (2020).
Huang, Y., Zhang, Y., Jia, L., Liu, C. & Xu, F. Circular RNA ABCB10 promotes tumor progression and correlates with pejorative prognosis in clear cell renal cell carcinoma. Int. J. Biol. Markers 34, 176–183 (2019).
Wang, Q. et al. Identification of METTL14 in kidney renal clear cell carcinoma using bioinformatics analysis. Dis. Markers 2019, 5648783 (2019).
Jeyaraman, S., Hanif, E. A. M., Ab Mutalib, N. S., Jamal, R. & Abu, N. Circular RNAs: potential regulators of treatment resistance in human cancers. Front. Genet. 10, 1369 (2019).
Yan, L., Liu, G., Cao, H., Zhang, H. & Shao, F. Hsa_circ_0035483 sponges hsa-miR-335 to promote the gemcitabine-resistance of human renal cancer cells by autophagy regulation. Biochem. Biophys. Res. Commun. 519, 172–178 (2019).
Li, C. M. et al. Circular RNA expression profiles in cisplatin-induced acute kidney injury in mice. Epigenomics. 11, 1191–1207 (2019).
He, N. et al. Analysis of circular RNA expression profile in HEK 293T cells exposed to ionizing radiation. Dose Response 17, 1559325819837795 (2019).
Holdt, L. M., Kohlmaier, A. & Teupser, D. Circular RNAs as therapeutic agents and targets. Front. Physiol. 9, 1262 (2018).
Zhang, M. & Xin, Y. Circular RNAs: a new frontier for cancer diagnosis and therapy. J. Hematol. Oncol. 11, 21 (2018).
Du, W. W. et al. Induction of tumor apoptosis through a circular RNA enhancing Foxo3 activity. Cell Death Differ. 24, 357–370 (2017).
Author information
Authors and Affiliations
Contributions
All authors wrote the article. J.M.L. researched the data and reviewed or edited the manuscript before submission. Correspondence regarding this Review should be sent to J.M.L. at johan.lorenzen@uzh.ch or j.m.lorenzen@gmail.com.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information
Nature Reviews Nephrology thanks R.-U. Müller, who co-reviewed with M. Ignarski, X.-M. Meng and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Intergenic
-
Region between two protein-coding genes.
- Intragenic
-
Region within a gene.
- Intronic
-
Intron region of a protein-coding gene.
- Alu elements
-
Short stretches of DNA that contain an abundance of transposable elements.
- Rolling circle amplification
-
An isothermal enzymatic process in which a short DNA or RNA primer is amplified to form a long single-stranded DNA or RNA using a circular DNA template and special DNA or RNA polymerases.
- Pyroptosis
-
A highly inflammatory form of lytic programmed cell death that occurs most frequently upon infection with intracellular pathogens and is likely to form part of the antimicrobial response.
- Enriched terms
-
Gene ontology term enrichment is a technique for interpreting sets of genes that makes use of the gene ontology system of classification, in which genes are assigned to a set of predefined bins on the basis of their functional characteristics.
- Endothelial to mesenchymal transition
-
(EndoMT). A process in which an endothelial cell undergoes a series of molecular events that lead to a change in phenotype towards a mesenchymal cell such as a myofibroblast or smooth muscle cell.
- Aptamers
-
Oligonucleotide or peptide molecules that bind to a specific target molecule.
Rights and permissions
About this article
Cite this article
van Zonneveld, A.J., Kölling, M., Bijkerk, R. et al. Circular RNAs in kidney disease and cancer. Nat Rev Nephrol 17, 814–826 (2021). https://doi.org/10.1038/s41581-021-00465-9
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41581-021-00465-9
This article is cited by
-
Circular RNA hsa_circ_0005519 contributes to acute kidney injury via sponging microRNA-98-5p
BMC Nephrology (2024)
-
EIF4A3-mediated biogenesis of circSTX6 promotes bladder cancer metastasis and cisplatin resistance
Journal of Experimental & Clinical Cancer Research (2024)
-
The role of circular RNA during the urological cancer metastasis: exploring regulatory mechanisms and potential therapeutic targets
Cancer and Metastasis Reviews (2024)
-
Extracellular vesicle-circEHD2 promotes the progression of renal cell carcinoma by activating cancer-associated fibroblasts
Molecular Cancer (2023)
-
Circular RNAs in renal cell carcinoma: from mechanistic to clinical perspective
Cancer Cell International (2023)