Abstract
Objective
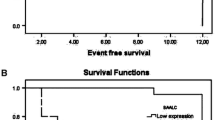
To investigate the expression and clinical significance of Bridging INtegrator 1 (BIN1) and cyclin D2 (CCND2) in newly diagnosed cytogenetically heterogenous adult acute myeloid leukemia patients. Methods Real-time quantitative PCR (RQ-PCR) was used to detect the expression of BIN1 and CCND2 genes in 49 newly diagnosed adult patients with AML, and their clinical significance was analyzed. Results BIN1 and CCND2 genes are highly expressed in patients with AML, which suggest their potential as molecular markers. A statistical significant correlation was found between BIN1 expression with bone marrow blasts (P = 0.012), CD34 expression (P = 0.026), NLR (P = 0.019) and Karyotype risk (P = 0.005). There was a statistically significant correlation between CCND2 and age (P = 0.001), NLR (P = 0.046) Karyotype risk (P = 0.033). The expression level of BIN1 was related to early treatment response (CR or not) (P = 0.043). CCND2 expression was significantly correlated with overall survival (P = 0.013), but not with CR (P = 0.731). Conclusion BIN1 and CCND2 genes are highly expressed in patients with AML, suggesting the potential of these two genes as molecular markers. Moreover, high levels of CCND2 are associated with better prognosis, suggesting the possibility of these two genes as prognostic markers.




Similar content being viewed by others
References
Siegel RL, Miller KD (2019) Jemal A (2019) Cancer statistics. CA Cancer J Clin 69(1):7–34. https://doi.org/10.3322/caac.21551
Green CL, Koo KK, Hills RK et al (2010) Prognostic significance of CEBPA mutations in a large cohort of younger adult patients with acute myeloid leukemia: impact of double CEBPA mutations and the interaction with FLT3 and NPM1 mutations. J Clin Oncol 28(16):2739–2747. https://doi.org/10.1200/jco.2009.26.2501
Pyndiah S, Tanida S, Ahmed KM et al (2011) c-MYC suppresses BIN1 to release poly (ADP-ribose) polymerase 1: a mechanism by which cancer cells acquire cisplatin resistance. Sci Signal 4(166):ra19. https://doi.org/10.1126/scisignal.2001556
Elliott K, Ge K, Du W et al (2000) The c-Myc-interacting adaptor protein Bin1 activates a caspase-independent cell death program. Oncogene 19(41):4669–4684. https://doi.org/10.1038/sj.onc.1203681
Muller AJ, Baker JF, DuHadaway JB et al (2003) Targeted disruption of the murine Bin1/Amphiphysin II gene does not disable endocytosis but results in embryonic cardiomyopathy with aberrant myofibril formation. Mol Cell Biol 23(12):4295–4306. https://doi.org/10.1128/MCB.23.12.4295-4306.2003
Esmailzadeh S, Huang Y, Su MW et al (2015) BIN1 tumor suppressor regulates Fas/Fas ligand-mediated apoptosis through c-FLIP in cutaneous T-cell lymphoma. Leukemia 29(6):1402–1413. https://doi.org/10.1038/leu.2015.9
Wang X, Wang J, Jia Y et al (2017) Methylation decreases the Bin1 tumor suppressor in ESCC and restoration by decitabine inhibits the epithelial mesenchymal transition. Oncotarget 8(12):19661–19673. https://doi.org/10.18632/oncotarget.14914
Jia Y, Wang H, Wang Y et al (2015) Low expression of Bin1, along with high expression of IDO in tumor tissue and draining lymph nodes, are predictors of poor prognosis for esophageal squamous cell cancer patients. Int J Cancer 137(5):1095–1106. https://doi.org/10.1002/ijc.29481
Wang J, Jia Y, Zhao S et al (2017) BIN1 reverses PD-L1-mediated immune escape by inactivating the c-MYC and EGFR/MAPK signaling pathways in non-small cell lung cancer. Oncogene 36(45):6235–6243. https://doi.org/10.1038/onc.2017.217
Ge Y, Schuster MB, Pundhir S et al (2019) The splicing factor RBM25 controls MYC activity in acute myeloid leukemia. Nat Commun 10(1):172. https://doi.org/10.1038/s41467-018-08076-y
Zhu C, Shao P, Bao M et al (2014) miR-154 inhibits prostate cancer cell proliferation by targeting CCND2. Urol Oncol 32(1):31.e9-31.e16. https://doi.org/10.1016/j.urolonc.2012.11.013
Zhou J, Tian Y, Li J et al (2013) miR-206 is down-regulated in breast cancer and inhibits cell proliferation through the up-regulation of cyclinD2. Biochem Biophys Res Commun 433(2):207–212. https://doi.org/10.1016/j.bbrc.2013.02.084
Du X, Lin LI, Zhang L et al (2015) microRNA-195 inhibits the proliferation, migration and invasion of cervical cancer cells via the inhibition of CCND2 and MYB expression. Oncol Lett 10(4):2639–2643. https://doi.org/10.3892/ol.2015.3541
Faber ZJ, Chen X, Gedman AL et al (2016) The genomic landscape of core-binding factor acute myeloid leukemias. Nat Genet 48(12):1551–1556. https://doi.org/10.1038/ng.3709
Eisfeld AK, Kohlschmidt J, Schwind S et al (2017) Mutations in the CCND1 and CCND2 genes are frequent events in adult patients with t(8;21)(q22;q22) acute myeloid leukemia. Leukemia 31(6):1278–1285. https://doi.org/10.1038/leu.2016.332
Martinez-Soria N, McKenzie L, Draper J et al (2018) The oncogenic transcription factor RUNX1/ETO corrupts cell cycle regulation to drive leukemic transformation. Cancer Cell 34(4):626-642.e8. https://doi.org/10.1016/j.ccell.2018.08.015
Liu W, Yi JM, Liu Y et al (2021) CDK6 Is a potential prognostic biomarker in acute myeloid leukemia. Front Genet 11:600227. https://doi.org/10.3389/fgene.2020.600227
Raeisi Shahraki H, Bemani P, Jalali M (2017) Classification of bladder cancer patients via penalized linear discriminant analysis. Asian Pac J Cancer Prev 18(5):1453–1457. https://doi.org/10.22034/APJCP.2017.18.5.1453
Vincenzi B, Stacchiotti S, Collini P et al (2017) Human equilibrative nucleoside transporter 1 gene expression is associated with gemcitabine efficacy in advanced leiomyosarcoma and angiosarcoma. Br J Cancer 117(3):340–346. https://doi.org/10.1038/bjc.2017.187
Pan K, Liang XT, Zhang HK et al (2012) Characterization of bridging integrator 1 (BIN1) as a potential tumor suppressor and prognostic marker in hepatocellular carcinoma. Mol Med 18(1):507–518. https://doi.org/10.2119/molmed.2011.00319
Tanida S, Mizoshita T, Ozeki K et al (2012) Mechanisms of cisplatin-induced apoptosis and of cisplatin sensitivity: potential of BIN1 to act as a potent predictor of cisplatin sensitivity in gastric cancer treatment. Int J Surg Oncol 2012:862879. https://doi.org/10.1155/2012/862879
Sun CC, Zhang L, Li G et al (2017) The lncRNA PDIA3P interacts with miR-185-5p to modulate oral squamous cell carcinoma progression by targeting cyclin D2. Mol Ther Nucleic Acids 9:100–110. https://doi.org/10.1016/j.omtn.2017.08.015
Wang Y, Xue J, Kuang H et al (2017) microRNA-1297 inhibits the growth and metastasis of colorectal cancer by suppressing cyclin D2 expression. DNA Cell Biol 36(11):991–999. https://doi.org/10.1089/dna.2017.3829
Takano Y, Kato Y, van Diest PJ et al (2000) Cyclin D2 overexpression and lack of p27 correlate positively and cyclin E inversely with a poor prognosis in gastric cancer cases. Am J Pathol 156(2):585–594. https://doi.org/10.1016/S0002-9440(10)64763-3
Sicinski P, Donaher JL, Geng Y et al (1996) Cyclin D2 is an FSH-responsive gene involved in gonadal cell proliferation and oncogenesis. Nature 384(6608):470–474. https://doi.org/10.1038/384470a0
Chen BB, Glasser JR, Coon TA et al (2012) F-box protein FBXL2 targets cyclin D2 for ubiquitination and degradation to inhibit leukemic cell proliferation. Blood 119(13):3132–3141. https://doi.org/10.1182/blood-2011-06-358911
Swart LE, Heidenreich O (2021) The RUNX1/RUNX1T1 network: translating insights into therapeutic options. Exp Hematol 94:1–10. https://doi.org/10.1016/j.exphem.2020.11.005
Hans CP, Weisenburger DD, Greiner TC et al (2004) Confirmation of the molecular classification of diffuse large B-cell lymphoma by immunohistochemistry using a tissue microarray. Blood 103(1):275–282. https://doi.org/10.1182/blood-2003-05-1545
Mermelshtein A, Gerson A, Walfisch S et al (2005) Expression of D-type cyclins in colon cancer and in cell lines from colon carcinomas. Br J Cancer 93(3):338–345. https://doi.org/10.1038/sj.bjc.6602709
He X, Chen SY, Yang Z et al (2018) miR-4317 suppresses non-small cell lung cancer (NSCLC) by targeting fibroblast growth factor 9 (FGF9) and cyclin D2 (CCND2). J Exp Clin Cancer Res 37(1):230. https://doi.org/10.1186/s13046-018-0882-4
Ko E, Kim Y, Park SE et al (2012) Reduced expression of cyclin D2 is associated with poor recurrence-free survival independent of cyclin D1 in stage III non-small cell lung cancer. Lung Cancer 77(2):401–406. https://doi.org/10.1016/j.lungcan.2012.03.027
Sarkar R, Hunter IA, Rajaganeshan R et al (2010) Expression of cyclin D2 is an independent predictor of the development of hepatic metastasis in colorectal cancer. Colorectal Dis 12(4):316–323. https://doi.org/10.1111/j.1463-1318.2009.01829.x
Matsubayashi H, Sato N, Fukushima N et al (2003) Methylation of cyclin D2 is observed frequently in pancreatic cancer but is also an age-related phenomenon in gastrointestinal tissues. Clin Cancer Res 9(4):1446–1452
Evron E, Umbricht CB, Korz D et al (2001) Loss of cyclin D2 expression in the majority of breast cancers is associated with promoter hypermethylation. Cancer Res 61(6):2782–2787
Ahmadzada T, Lee K, Clarke C et al (2019) High BIN1 expression has a favorable prognosis in malignant pleural mesothelioma and is associated with tumor infiltrating lymphocytes. Lung Cancer 130:35–41. https://doi.org/10.1016/j.lungcan.2019.02.005
Fang X, Chen C, Xia F et al (2016) CD274 promotes cell cycle entry of leukemia-initiating cells through JNK/Cyclin D2 signaling. J Hematol Oncol 9(1):124. https://doi.org/10.1186/s13045-016-0350-6
Acknowledgements
This study was supported by Sichuan Department of Science and Technology (Grant No. 20YYJC0940) and Lu zhou people's government- Southwest Medical University joint Project (Grant No. 2017LZXNYD-Z06)
Author information
Authors and Affiliations
Contributions
For every author, his or her contribution to the manuscript needs to be provided using the following categories: CONCEPTION: Chunlan Huang, Xinwen Zhang, Hao Xiong, INTERPRETATION OR ANALYSIS OF DATA: Xinwen Zhang, Hao Xiong, Xiaomin Chen, Yang Liu, Jialin Duan. PREPARATION OF THE MANUSCRIPT: Xinwen Zhang, Hao Xiong. REVISION FOR IMPORTANT INTELLECTUAL CONTENT: Xinwen Zhang, Hao Xiong, Xiaomin Chen, Yang Liu, Jialin Duan. SUPERVISION: Chunlan Huang.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interests.
Ethical Approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional committee.
Informed Consent
Informed consent was obtained from all individual participants included in the study.
Human and Animal Participants
This study involves blood and bone marrow aspirates from AML patients. Ethical approval was obtained from institutional ethics committee (AF-V4-22.0) to conduct the study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Zhang, X., Xiong, H., Duan, J. et al. The Prognostic Significance of the BIN1 and CCND2 Gene in Adult Patients with Acute Myeloid Leukemia. Indian J Hematol Blood Transfus 38, 481–491 (2022). https://doi.org/10.1007/s12288-021-01479-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12288-021-01479-w