Published online Jul 26, 2021. doi: 10.4252/wjsc.v13.i7.934
Peer-review started: February 28, 2021
First decision: April 19, 2021
Revised: May 5, 2021
Accepted: July 9, 2021
Article in press: July 9, 2021
Published online: July 26, 2021
The classical cancer stem cell (CSCs) theory proposed the existence of a rare but constant subpopulation of CSCs. In this model cancer cells are organized hierarchically and are responsible for tumor resistance and tumor relapse. Thus, eliminating CSCs will eventually lead to cure of cancer. This simplistic model has been challenged by experimental data. In 2010 we proposed a novel and controversial alternative model of CSC biology (the Stemness Phenotype Model, SPM). The SPM proposed a non-hierarchical model of cancer biology in which there is no specific subpopulation of CSCs in tumors. Instead, cancer cells are highly plastic in term of stemness and CSCs and non-CSCs can interconvert into each other depending on the microenvironment. This model predicts the existence of cancer cells ranging from a pure CSC phenotype to pure non-CSC phenotype and that survival of a single cell can originate a new tumor. During the past 10 years, a plethora of experimental evidence in a variety of cancer types has shown that cancer cells are indeed extremely plastic and able to interconvert into cells with different stemness phenotype. In this review we will (1) briefly describe the cumulative evidence from our laboratory and others supporting the SPM; (2) the implications of the SPM in translational oncology; and (3) discuss potential strategies to develop more effective therapeutic regimens for cancer treatment.
Core Tip: The classical cancer stem cell theory proposed the existence of a rare but constant subpopulation of cancer stem cells. This review article briefly describes the cumulative evidence supporting alternative models of cancer stem cells, their implications in translational oncology and, discuss the potential strategies to develop more effective and less toxic sequential multistep-based therapeutic regimens for cancer treatment.
- Citation: Kaushik V, Kulkarni Y, Felix K, Azad N, Iyer AKV, Yakisich JS. Alternative models of cancer stem cells: The stemness phenotype model, 10 years later. World J Stem Cells 2021; 13(7): 934-943
- URL: https://www.wjgnet.com/1948-0210/full/v13/i7/934.htm
- DOI: https://dx.doi.org/10.4252/wjsc.v13.i7.934
The biological properties of cancer cells have profound implications for all areas of oncology research ranging from preclinical studies to advanced clinical trials. It is not a surprise that numerous conceptual models of cancer cell biology have been proposed with the ultimate goal to develop effective therapies that not only extend survival but lead to a definitive cure. In the past decades the discovery of the potential abilities in self-renewal and differentiation of normal stem cells has opened a new horizon in medicine and important concepts were extrapolated to neoplastic cells. Although the concept of cancer stem cell (CSC) is not new since the general idea of tumors driven by a subset of cells endowed with stem-like properties was postulated by Rudolf Virchow in 1855[1] it has gained tremendous momentum after the isolation of putative cancer stem-like cells (CS-LCs) in a variety of cancer types including brain tumors[2-5], breast cancer[6-9], colon[10], hepatic[11], pancreatic[12], thyroid[13,14], bladder[15,16], cervical[17-20], ovarian[21-24], urothelial[25-28], renal[29-31] and chordoma[32,33]. In general, putative CS-LCs were isolated from every type of fresh tumor specimens and cancer cell lines. These discoveries quickly led to a new paradigm, the so called “Cancer Stem Cell Theory” (CSCT), that is fundamentally not completely different from the original models proposed by Virchow and his contemporaries[1]. In this model there is a hierarchical organization where a subset of CSCs can irreversibly differentiate into all types of non-CSCs. Prior to the modern CSCT, that started with the first isolation of putative CS-LCs in 1997[34] the clonal stochastic model (cSM) postulated in 1976[35] was popular among oncologists. The cSM proposed that all transformed cells in the tumor have carcinogenic potential and are able to proliferate and produce the same cells. The cSM is a non-hierachical model. From the clinical point of view, according to the cSM, to cure cancer all cancer cells should be eliminated since any cancer cell is potentially tumorigenic. On the contrary, according to the modern CSCT, to cure cancer or at least to obtain a significant outcome, it should be enough to eliminate only the rare subpopulation of CSCs. The idea of a rare subpopulation of CSCs as driving element in cancer development, evolution and heterogeneity, has overridden the previous cSM model[7] and catapulted research of therapeutic strategies based on CSCs targeting, such as the targeting of CSC niche, CSC signaling pathways, and CSC mitochondria and, metabolism[7,36-38].
Although the modern CSCT was an attractive concept it was found soon to be insufficient to reconcile experimental findings with a hierarchical rigid model. As a consequence several alternative plasticity models of CSCs such as “Stemness Phenotype Model”[39], the “complex system model”[40], the “Dynamic CSC model”[41], and the “Dedifferentiation model”[42] were proposed as early as 2010. These models share some similarities, but a detailed comparison is beyond the scope of this article. For review see Cruz et al[43]. The aim of this short review is to update and highlight key predictions of the Stemness Phenotype Model (SPM).
In short, the SPM was originally proposed as a “One compartment model” where there is only one cancer cell type. These cells are cells with different stemness phenotype due to random biological variation. The stemness depends on the microenvironment where the cells grow and can range from a phenotype resembling a non-CSC to a pure CSC. In other words, the SPM proposed that there are no true different subpopulations of CSCs and non-CSCs but a single cell type that can interconvert into each other depending on the microenvironmental conditions. An immediate prediction of this model is that there are cells having “intermediate phenotypes” between both extreme phenotypes[39]. Other key prediction of the SPM is that the survival of a single cell might induce tumor relapse and therefore any effective therapy will must be able to eliminate 100% of cancer cells at once in order to prevent regrowth[43].
In the SPM microenvironmentally-driven interconversion between CSCs and non-CSCs is a key process that explains the characteristic found in tumors such as the existence of intratumoral heterogeneity and chemoresistance. Evidence of intratumoral heterogeneity due to interconversion between cancer cell phenotypes was likely observed long before the first isolation of putative CSCs. For instance, in 1987 it was reported that several different cell phenotypes coexist in the human breast cancer cell line MCF7[44]. Similarly, it was known by 1998 that the human lung carcinoma cell line DLKP contains 3 distinct subpopulations and that two of them can interconvert to the third one[45]. At present, the findings of these two examples can easily be explained by interconversion but at that time the concept of stemness was not common in the literature. Definitive evidence of microenvironmentally-driven interconversion between CSCs and non-CSCs phenotypes were already available after the isolation of putative CSCs (characterized by stemness markers) when it was found that cells that were considered non-CSCs could interconvert into CSCs. For instance, (1) some CD44− Du145 prostate cancer cells (100% purity) could give rise to CD44+ cells in culture[46]; and (2) non-SP MCF7 breast cancer cells when recultured after being sorted contained SP cells indicating that the non-SP fraction gave rise to a new SP subpopulation[47]. A direct conversion from a non-CSC phenotype to a CSC phenotype was demonstrated in breast and prostate cancer cells when it was observed that exposure to conditioned media stimulated non-CSCs to become CSCs and that IL6 was enough to drive this conversion[48]. Other examples include the ability of some mature leukemia cells to de-differentiate and reacquire clonogenic and leukemogenic properties[49] and the de-differentiation of glioma cells to glioma stem-like cells by therapeutic stress[50]. Additional evidence of non-CSCs conversion into CSCs were found in osteosarcoma[51], lung[52], pancreatic[53], colon[54] and breast cancers[55]. In vitro data from our lab and others demonstrated that phenotypic changes due to changes in culture conditions are rapid and reversible[56,57]. For instance, cancer cells can become (within three days) highly resistant to conventional anticancer drugs when switched from anchorage-dependent (adherent cells) culture conditions into anchorage-independent (floating cells) culture conditions. Chemosensitivity was quickly restored (within three days) when floating cells were cultured back as adherent cells. Under these conditions, a reversible change in the expression of proteins from multiple pathways was observed demonstrating complex and quick phenotypic adaptations to changing environment[56,57].
The SPM predicts the existence of multiple subpopulations of cancer cells ranging from a “pure non-CSC phenotype” to a “pure CSC phenotype”. This prediction was confirmed in the non-small cell lung adenocarcinoma (NSCLA) cell lines A549 and H441. It was found that NSCLA cells contain multiple, interconvertible, phenotypically distinct subpopulations (e.g., non-SP, SP, CD133pos and ALDHhigh) that exhibit distinct self-renewal and metastatic gene expression patterns[52].
These findings clearly demonstrated that cancer cells are actually extremely plastic and that microenvironmental conditions can influence and drive the bidirectional interconversion between CSCs and non-CSCs phenotypes. Recently several theoretical multi-phenotypic models that include, interconversion and cellular plasticity has been useful in predicting and validating this new paradigm[58-60].
The ability of any given cancer cell regardless of its phenotype to reconstitute in vivo the complex intratumoral heterogeneity of any cancer is the ultimate prediction of the SPM. In vivo evidence suggesting that any cancer cell is potentially tumorigenic were available long before any alternative model of CSCs were published. Perhaps the most convincing data was published in 2007 demonstrating that each of the 67 single C6 glioma (Including CD133-) cells plated per miniwell was able to generate a clone and subclones, which subsequently gave rise to a xenograft glioma in the BALB/C-nude mouse[61]. Recently, it has been reported that all 16 subpopulations of highly heterogeneous GBM cultures carry stem cell properties in vitro. These cells undergo stochastic state transitions, they showed reversible phenotypic adaptation in vivo and they all formed tumors. More importantly, the authors showed that the phenotypic heterogeneity could also be recreated by single cells of different phenotypic profiles[62].
Mathematical models of cancer biology are providing insight of strategies for cancer elimination. Simple mathematical models considering two populations of cells: CSCs, which can divide indefinitely, and differentiated cancer cells, which do not divide and have a limited lifespan predict that neither inhibition of CSCs proliferation alone nor stimulation of CSCs differentiation is sufficient for cancer cure[63].
Mathematic modelling of in vitro growth of heterogeneous cell cultures in the presence of interconversion from differentiated cancer cells to CSCs also demonstrated that by targeting only the CSCs subpopulation will not be enough to eradicate cancer and that the chemotherapeutic elimination of in vitro cultures of heterogeneous cancer cells will be effective only if it targets all cancer cell types[64]. From the clinical point of view, the SPM seems to bring back the field of cancer treatment research to the early days of the cSM. The overall clinical implications of both the SPM and the cSM are essentially the same: they both predict that to cure cancer all cancer cells should be eliminated. However, these two models are conceptually very different and, it can be predicted that to achieve complete elimination of all cancer cells (if we ever achieve that goal) it will require a different approach. It is likely then that a successful chemotherapy regime will require numerous anticancer therapies, each of them targeting a “spectrum” of cancer cell subpopulations that in turn can create serious toxicity issues. The next big challenge in the oncotherapy field will be to develop a safe (low or non-toxic) therapeutic regime that can be administered simultaneously to deplete all cancer cells at once.
In complex, highly heterogeneous tumors the eradication of all cancer cells at once will likely require high doses of anticancer agents +/- radiation/immunotherapy that will severely limit its practical application due to toxicity issues. One alternative to circumvent this problem is to administer them sequentially. Sequential cancer treatment with chemotherapy followed by radiotherapy + high dose chemotherapy followed by autologous peripheral blood stem cell transplantation (APBSCT) has been employed with relatively good outcomes in several cancers such as mantle cell lymphoma[65] and relapsed/refractory acute myeloid leukemia[66]. Sequential multimodalities regimes are being increasingly utilized to treat patients carrying different types of cancers such as gastric cancer[67], pancreatic cancer[68], leukemia[69], non-small cell lung cancer[70] and, breast cancer[71]. Sequential anthracycline- and taxane-based neoadjuvant chemotherapy represents the standard therapeutic approach for the majority of patients with early-stage triple negative breast cancer[72]. Novel sequential treatment are also currently investigated at the preclinical level[73]. We have demonstrated that a first step treatment with Hydroxyurea left few DBTRG.05MG glioma cells arrested in a senescent-like state. In the second step, salinomycin at low concentration eliminated 100% of these senescent-like cells[74]. These cells can grow in suspension as neurospheres, in which the Hedgehog pathway is activated[75]. This in vitro example of sequential chemotherapy suggests that it would be possible to eliminate all cancer cells at once with lower, and therefore less, toxic concentrations.
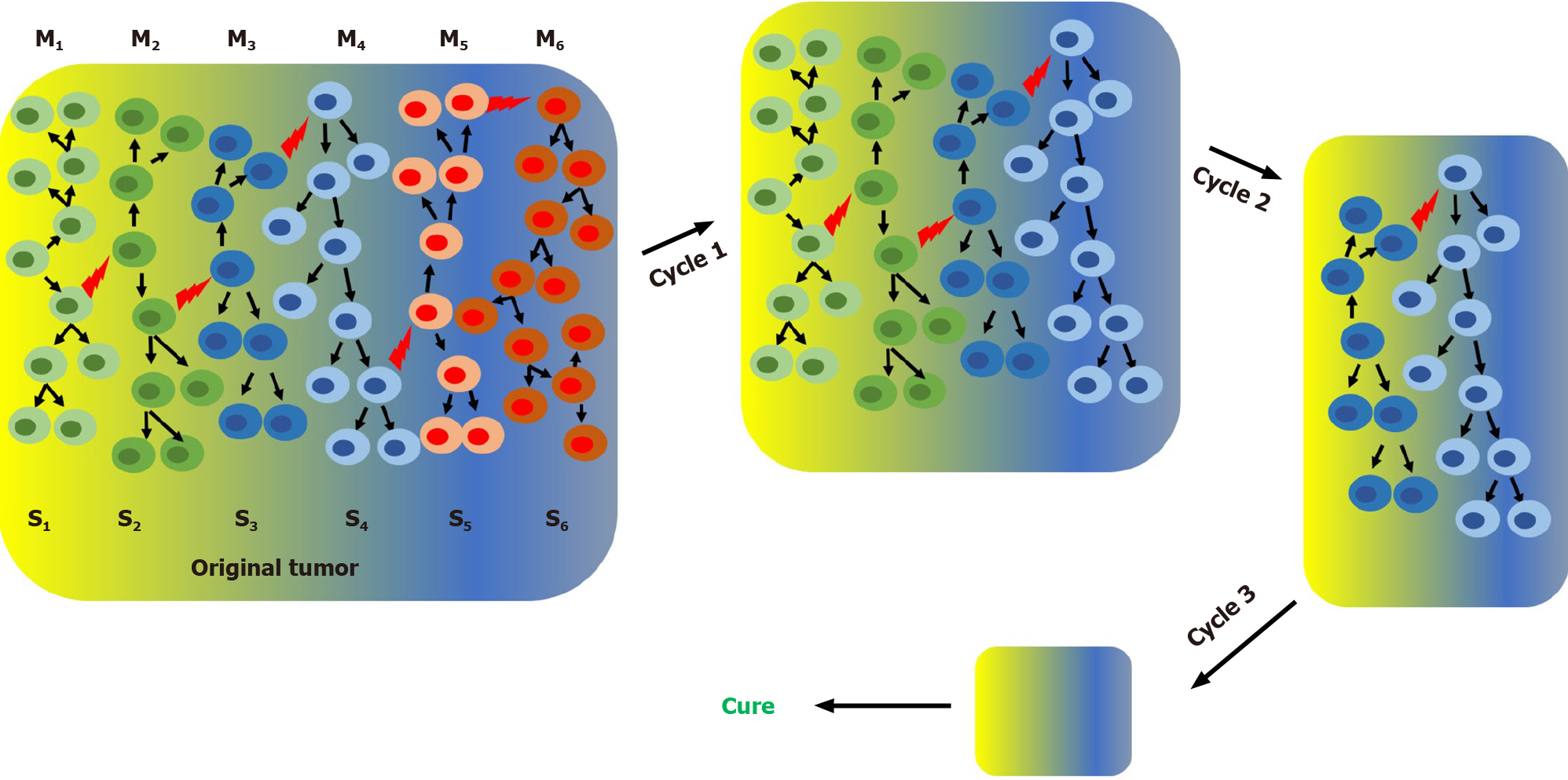
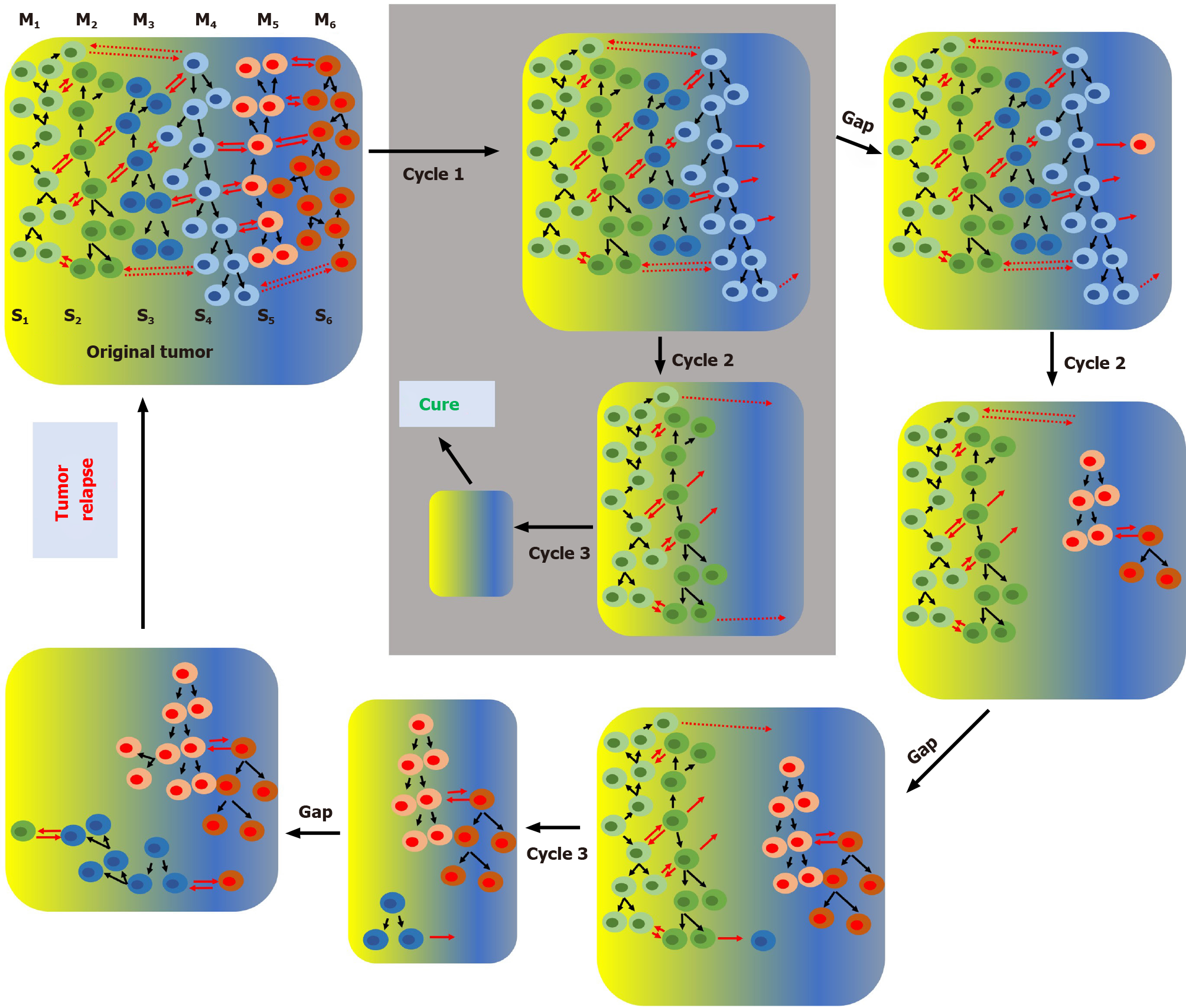
In the cSM, tumor heterogeneity appears as consequence of random genetic changes concomitant with clonal selection. At some point during its development a tumor may have different genetically defined subpopulations growing in specific microenvironments. Due to the irreversible nature of genetic mutations, specific subpopulations can be permanently eliminated with specific anticancer treatments. For instance, sequential chemotherapy steps with different anticancer drugs can potentially eliminate one or few subpopulations per step which will eventually lead to a cure when the last subpopulation is eliminated (Figure 1). It is important to point out that, due to the high genetic instability of cancer cells, any time gap between steps increases the chances of generating new genetic clones (new cancer cell subpopulations) and thus increases the chances of tumor relapse. According to the SPM any time gap between steps increases the chances of regenerating the cancer cell subpopulation(s) eliminated in previous steps by interconversion. We have demonstrated in vitro that cancer cells are extremely plastic and they can undergo cycles of phenotypic changes within few days[57]. According to the SPM, multistep treatment regimes may only work if there is no time gap or just gap between steps or if the interconversion process is inhibited (Figure 2).
The SPM does not exclude the coexistence of other mechanisms contributing to intratumoral heterogeneity. For instance, in addition to microenvironmentally-driven interconversion between CSCs and non-CSCs, genetic mutations due to genomic instability of cancer cells may create new subpopulations of CSCs in the same tumor[43]. Key concepts from other models not only expand our knowledge of cancer biology but can be useful in designing a curative treatment. For instance, in addition to blocking interconversion, it could be helpful to prevent the generation of new clones originated by stochastic genetic mutations. One of the hallmark of cancer cells is “avoidance of apoptosis” following, for instance, DNA damage[76]. In this context, drugs that promote apoptosis induced by DNA-damage such as PARP alone or in combination with ATR inhibitors[77,78], can potentially reduce the generation of new genetic clones.
During the last decade the SPM and similar alternative models of cancer biology have expanded our understanding of cancer biology and new therapeutic targets and biological processes have been identified. For instance, targeting key factors involved in the process of interconversion opens the opportunity to block the conversion of a non-CSCs phenotype into a CSC phenotype and thus reducing chemoresistance and tumor relapse. The SPM, initially proposed for gliomas in a conceptual review article in 2010[39] was quickly extrapolated to other types of tumors[43]. During the last ten years, extensive experimental evidence was published supporting the notion that microenvironmentally-driven interconversion between CSCs and non-CSCs is a key process leading to intratumoral heterogeneity, that in turn is responsible for chemoresistance and tumor relapse. Additionally, key predictions of the SPM – the ability of any given cancer cell to reconstitute in vivo the complex intratumoral heterogeneity has been demonstrated experimentally in gliomas. The SPM has been demonstrated to be a useful working model of cancer biology that should be taken in consideration when developing new cancer treatment modalities. In addition, the SPM is a model of cancer biology (at the cellular level) that does not necessarily exclude key concepts from other cancer models and therefore has the potential to integrate them into more complex tumoral (at the tissue level) models that can be experimentally tested for developing novel treatments.
Manuscript source: Invited manuscript
Specialty type: Cell and tissue engineering
Country/Territory of origin: United States
Peer-review report’s scientific quality classification
Grade A (Excellent): 0
Grade B (Very good): B
Grade C (Good): 0
Grade D (Fair): 0
Grade E (Poor): 0
P-Reviewer: Cao ZF S-Editor: Gao CC L-Editor: A P-Editor: Liu JH
| 1. | Raggi C, Mousa HS, Correnti M, Sica A, Invernizzi P. Cancer stem cells and tumor-associated macrophages: a roadmap for multitargeting strategies. Oncogene. 2016;35:671-682. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 87] [Cited by in F6Publishing: 110] [Article Influence: 12.2] [Reference Citation Analysis (0)] |
| 2. | Guan R, Zhang X, Guo M. Glioblastoma stem cells and Wnt signaling pathway: molecular mechanisms and therapeutic targets. Chin Neurosurg J. 2020;6:25. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 14] [Cited by in F6Publishing: 22] [Article Influence: 5.5] [Reference Citation Analysis (0)] |
| 3. | Liu HL, Wang YN, Feng SY. Brain tumors: Cancer stem-like cells interact with tumor microenvironment. World J Stem Cells. 2020;12:1439-1454. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 3] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 4. | Ryskalin L, Gaglione A, Limanaqi F, Biagioni F, Familiari P, Frati A, Esposito V, Fornai F. The Autophagy Status of Cancer Stem Cells in Gliobastoma Multiforme: From Cancer Promotion to Therapeutic Strategies. Int J Mol Sci. 2019;20. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 39] [Article Influence: 7.8] [Reference Citation Analysis (0)] |
| 5. | Suvà ML, Tirosh I. The Glioma Stem Cell Model in the Era of Single-Cell Genomics. Cancer Cell. 2020;37:630-636. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 99] [Cited by in F6Publishing: 129] [Article Influence: 32.3] [Reference Citation Analysis (0)] |
| 6. | Dahn ML, Marcato P. Targeting the Roots of Recurrence: New Strategies for Eliminating Therapy-Resistant Breast Cancer Stem Cells. Cancers (Basel). 2020;13. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 3] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 7. | Shan NL, Shin Y, Yang G, Furmanski P, Suh N. Breast cancer stem cells: A review of their characteristics and the agents that affect them. Mol Carcinog. 2021;60:73-100. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 26] [Article Influence: 8.7] [Reference Citation Analysis (0)] |
| 8. | Zhang X, Powell K, Li L. Breast Cancer Stem Cells: Biomarkers, Identification and Isolation Methods, Regulating Mechanisms, Cellular Origin, and Beyond. Cancers (Basel). 2020;12. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 54] [Cited by in F6Publishing: 38] [Article Influence: 9.5] [Reference Citation Analysis (0)] |
| 9. | Zheng Q, Zhang M, Zhou F, Zhang L, Meng X. The Breast Cancer Stem Cells Traits and Drug Resistance. Front Pharmacol. 2020;11:599965. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 17] [Cited by in F6Publishing: 17] [Article Influence: 5.7] [Reference Citation Analysis (0)] |
| 10. | Ricci-Vitiani L, Fabrizi E, Palio E, De Maria R. Colon cancer stem cells. J Mol Med (Berl). 2009;87:1097-1104. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 139] [Cited by in F6Publishing: 164] [Article Influence: 10.9] [Reference Citation Analysis (0)] |
| 11. | Shen Y, Cao D. Hepatocellular carcinoma stem cells: origins and roles in hepatocarcinogenesis and disease progression. Front Biosci (Elite Ed). 2012;4:1157-1169. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 13] [Article Influence: 1.1] [Reference Citation Analysis (0)] |
| 12. | Wang Z, Ahmad A, Li Y, Azmi AS, Miele L, Sarkar FH. Targeting notch to eradicate pancreatic cancer stem cells for cancer therapy. Anticancer Res. 2011;31:1105-1113. [PubMed] [Cited in This Article: ] |
| 13. | Thomas D, Friedman S, Lin RY. Thyroid stem cells: lessons from normal development and thyroid cancer. Endocr Relat Cancer. 2008;15:51-58. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 43] [Cited by in F6Publishing: 43] [Article Influence: 2.7] [Reference Citation Analysis (0)] |
| 14. | Zhang P, Zuo H, Ozaki T, Nakagomi N, Kakudo K. Cancer stem cell hypothesis in thyroid cancer. Pathol Int. 2006;56:485-489. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 56] [Cited by in F6Publishing: 60] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 15. | Abugomaa A, Elbadawy M, Yamawaki H, Usui T, Sasaki K. Emerging Roles of Cancer Stem Cells in Bladder Cancer Progression, Tumorigenesis, and Resistance to Chemotherapy: A Potential Therapeutic Target for Bladder Cancer. Cells. 2020;9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 42] [Article Influence: 10.5] [Reference Citation Analysis (0)] |
| 16. | Tran MN, Goodwin Jinesh G, McConkey DJ, Kamat AM. Bladder cancer stem cells. Curr Stem Cell Res Ther. 2010;5:387-395. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 29] [Cited by in F6Publishing: 31] [Article Influence: 2.4] [Reference Citation Analysis (0)] |
| 17. | Huang R, Rofstad EK. Cancer stem cells (CSCs), cervical CSCs and targeted therapies. Oncotarget. 2017;8:35351-35367. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 76] [Cited by in F6Publishing: 85] [Article Influence: 14.2] [Reference Citation Analysis (0)] |
| 18. | López J, Poitevin A, Mendoza-Martínez V, Pérez-Plasencia C, García-Carrancá A. Cancer-initiating cells derived from established cervical cell lines exhibit stem-cell markers and increased radioresistance. BMC Cancer. 2012;12:48. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 136] [Cited by in F6Publishing: 149] [Article Influence: 12.4] [Reference Citation Analysis (0)] |
| 19. | Mendoza-Almanza G, Ortíz-Sánchez E, Rocha-Zavaleta L, Rivas-Santiago C, Esparza-Ibarra E, Olmos J. Cervical cancer stem cells and other leading factors associated with cervical cancer development. Oncol Lett. 2019;18:3423-3432. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 22] [Article Influence: 4.4] [Reference Citation Analysis (0)] |
| 20. | Organista-Nava J, Gómez-Gómez Y, Garibay-Cerdenares OL, Leyva-Vázquez MA, Illades-Aguiar B. Cervical cancer stem cell-associated genes: Prognostic implications in cervical cancer. Oncol Lett. 2019;18:7-14. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 18] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 21. | Aguilar-Gallardo C, Rutledge EC, Martínez-Arroyo AM, Hidalgo JJ, Domingo S, Simón C. Overcoming challenges of ovarian cancer stem cells: novel therapeutic approaches. Stem Cell Rev Rep. 2012;8:994-1010. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 38] [Cited by in F6Publishing: 38] [Article Influence: 3.2] [Reference Citation Analysis (0)] |
| 22. | Ahmed N, Kadife E, Raza A, Short M, Jubinsky PT, Kannourakis G. Ovarian Cancer, Cancer Stem Cells and Current Treatment Strategies: A Potential Role of Magmas in the Current Treatment Methods. Cells. 2020;9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 23] [Cited by in F6Publishing: 27] [Article Influence: 6.8] [Reference Citation Analysis (0)] |
| 23. | Howard CM, Zgheib NB, Bush S 2nd, DeEulis T, Cortese A, Mollo A, Lirette ST, Denning K, Valluri J, Claudio PP. Clinical relevance of cancer stem cell chemotherapeutic assay for recurrent ovarian cancer. Transl Oncol. 2020;13:100860. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 3] [Article Influence: 0.8] [Reference Citation Analysis (0)] |
| 24. | Terraneo N, Jacob F, Dubrovska A, Grünberg J. Novel Therapeutic Strategies for Ovarian Cancer Stem Cells. Front Oncol. 2020;10:319. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 30] [Cited by in F6Publishing: 32] [Article Influence: 8.0] [Reference Citation Analysis (0)] |
| 25. | Fang D, Kitamura H. Cancer stem cells and epithelial-mesenchymal transition in urothelial carcinoma: Possible pathways and potential therapeutic approaches. Int J Urol. 2018;25:7-17. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 25] [Cited by in F6Publishing: 38] [Article Influence: 5.4] [Reference Citation Analysis (0)] |
| 26. | Garg M. Urothelial cancer stem cells and epithelial plasticity: current concepts and therapeutic implications in bladder cancer. Cancer Metastasis Rev. 2015;34:691-701. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 44] [Cited by in F6Publishing: 55] [Article Influence: 6.9] [Reference Citation Analysis (0)] |
| 27. | Ho PL, Kurtova A, Chan KS. Normal and neoplastic urothelial stem cells: getting to the root of the problem. Nat Rev Urol. 2012;9:583-594. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 143] [Cited by in F6Publishing: 150] [Article Influence: 12.5] [Reference Citation Analysis (0)] |
| 28. | Kripnerova M, Parmar HS, Pesta M, Kohoutova M, Kuncova J, Drbal K, Rajtmajerova M, Hatina J. Urothelial Cancer Stem Cell Heterogeneity. Adv Exp Med Biol. 2019;1139:127-151. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 5] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 29. | Fang P, Zhou L, Lim LY, Fu H, Yuan ZX, Lin J. Targeting Strategies for Renal Cancer Stem Cell Therapy. Curr Pharm Des. 2020;26:1964-1978. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 5] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 30. | Khan MI, Czarnecka AM, Helbrecht I, Bartnik E, Lian F, Szczylik C. Current approaches in identification and isolation of human renal cell carcinoma cancer stem cells. Stem Cell Res Ther. 2015;6:178. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 49] [Cited by in F6Publishing: 50] [Article Influence: 5.6] [Reference Citation Analysis (0)] |
| 31. | Liu Q, Gu J, Zhang E, He L, Yuan ZX. Targeted Delivery of Therapeutics to Urological Cancer Stem Cells. Curr Pharm Des. 2020;26:2038-2056. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4] [Cited by in F6Publishing: 4] [Article Influence: 1.0] [Reference Citation Analysis (0)] |
| 32. | Aydemir E, Bayrak OF, Sahin F, Atalay B, Kose GT, Ozen M, Sevli S, Dalan AB, Yalvac ME, Dogruluk T, Türe U. Characterization of cancer stem-like cells in chordoma. J Neurosurg. 2012;116:810-820. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 44] [Article Influence: 3.7] [Reference Citation Analysis (0)] |
| 33. | Tuysuz EC, Gulluoglu S, Yaltirik CK, Ozbey U, Kuskucu A, Çoban EA, Sahin F, Türe U, Bayrak OF. Distinctive role of dysregulated miRNAs in chordoma cancer stem-like cell maintenance. Exp Cell Res. 2019;380:9-19. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 7] [Article Influence: 1.4] [Reference Citation Analysis (0)] |
| 34. | Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3:730-737. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4851] [Cited by in F6Publishing: 4678] [Article Influence: 173.3] [Reference Citation Analysis (1)] |
| 35. | Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23-28. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 4428] [Cited by in F6Publishing: 4025] [Article Influence: 83.9] [Reference Citation Analysis (1)] |
| 36. | Duan H, Liu Y, Gao Z, Huang W. Recent advances in drug delivery systems for targeting cancer stem cells. Acta Pharm Sin B. 2021;11:55-70. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 99] [Cited by in F6Publishing: 102] [Article Influence: 34.0] [Reference Citation Analysis (0)] |
| 37. | Gao X, Dong QZ. Advance in metabolism and target therapy in breast cancer stem cells. World J Stem Cells. 2020;12:1295-1306. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 10] [Cited by in F6Publishing: 10] [Article Influence: 2.5] [Reference Citation Analysis (0)] |
| 38. | Mukha A, Dubrovska A. Metabolic Targeting of Cancer Stem Cells. Front Oncol. 2020;10:537930. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 14] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 39. | Cruz M, Siden Å, Tasat DR, Yakisich JS. Are all glioma cells cancer stem cells? J Cancer Sci Ther. 2010;2:100-106. [Cited in This Article: ] |
| 40. | Laks DR, Visnyei K, Kornblum HI. Brain tumor stem cells as therapeutic targets in models of glioma. Yonsei Med J. 2010;51:633-640. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 24] [Cited by in F6Publishing: 28] [Article Influence: 2.0] [Reference Citation Analysis (0)] |
| 41. | Vermeulen L, de Sousa e Melo F, Richel DJ, Medema JP. The developing cancer stem-cell model: clinical challenges and opportunities. Lancet Oncol. 2012;13:e83-e89. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 262] [Cited by in F6Publishing: 267] [Article Influence: 22.3] [Reference Citation Analysis (0)] |
| 42. | Li Y, Laterra J. Cancer stem cells: distinct entities or dynamically regulated phenotypes? Cancer Res. 2012;72:576-580. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 149] [Cited by in F6Publishing: 156] [Article Influence: 13.0] [Reference Citation Analysis (0)] |
| 43. | Cruz MH, Sidén A, Calaf GM, Delwar ZM, Yakisich JS. The stemness phenotype model. ISRN Oncol. 2012;2012:392647. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 13] [Cited by in F6Publishing: 22] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 44. | Resnicoff M, Medrano EE, Podhajcer OL, Bravo AI, Bover L, Mordoh J. Subpopulations of MCF7 cells separated by Percoll gradient centrifugation: a model to analyze the heterogeneity of human breast cancer. Proc Natl Acad Sci USA. 1987;84:7295-7299. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 57] [Cited by in F6Publishing: 59] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 45. | McBride S, Meleady P, Baird A, Dinsdale D, Clynes M. Human lung carcinoma cell line DLKP contains 3 distinct subpopulations with different growth and attachment properties. Tumour Biol. 1998;19:88-103. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 16] [Article Influence: 0.7] [Reference Citation Analysis (0)] |
| 46. | Patrawala L, Calhoun T, Schneider-Broussard R, Li H, Bhatia B, Tang S, Reilly JG, Chandra D, Zhou J, Claypool K, Coghlan L, Tang DG. Highly purified CD44+ prostate cancer cells from xenograft human tumors are enriched in tumorigenic and metastatic progenitor cells. Oncogene. 2006;25:1696-1708. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 727] [Cited by in F6Publishing: 709] [Article Influence: 39.4] [Reference Citation Analysis (0)] |
| 47. | Zhang Y, Piao B, Zhang Y, Hua B, Hou W, Xu W, Qi X, Zhu X, Pei Y, Lin H. Oxymatrine diminishes the side population and inhibits the expression of β-catenin in MCF-7 breast cancer cells. Med Oncol. 2011;28 Suppl 1:S99-107. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 52] [Cited by in F6Publishing: 50] [Article Influence: 3.6] [Reference Citation Analysis (0)] |
| 48. | Iliopoulos D, Hirsch HA, Wang G, Struhl K. Inducible formation of breast cancer stem cells and their dynamic equilibrium with non-stem cancer cells via IL6 secretion. Proc Natl Acad Sci USA. 2011;108:1397-1402. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 484] [Cited by in F6Publishing: 512] [Article Influence: 39.4] [Reference Citation Analysis (0)] |
| 49. | McKenzie MD, Ghisi M, Oxley EP, Ngo S, Cimmino L, Esnault C, Liu R, Salmon JM, Bell CC, Ahmed N, Erlichster M, Witkowski MT, Liu GJ, Chopin M, Dakic A, Simankowicz E, Pomilio G, Vu T, Krsmanovic P, Su S, Tian L, Baldwin TM, Zalcenstein DA, DiRago L, Wang S, Metcalf D, Johnstone RW, Croker BA, Lancaster GI, Murphy AJ, Naik SH, Nutt SL, Pospisil V, Schroeder T, Wall M, Dawson MA, Wei AH, de Thé H, Ritchie ME, Zuber J, Dickins RA. Interconversion between Tumorigenic and Differentiated States in Acute Myeloid Leukemia. Cell Stem Cell. 2019;25:258-272.e9. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 41] [Cited by in F6Publishing: 30] [Article Influence: 7.5] [Reference Citation Analysis (0)] |
| 50. | Lee G, Auffinger B, Guo D, Hasan T, Deheeger M, Tobias AL, Kim JY, Atashi F, Zhang L, Lesniak MS, James CD, Ahmed AU. Dedifferentiation of Glioma Cells to Glioma Stem-like Cells By Therapeutic Stress-induced HIF Signaling in the Recurrent GBM Model. Mol Cancer Ther. 2016;15:3064-3076. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 69] [Cited by in F6Publishing: 79] [Article Influence: 9.9] [Reference Citation Analysis (0)] |
| 51. | Zhang H, Wu H, Zheng J, Yu P, Xu L, Jiang P, Gao J, Wang H, Zhang Y. Transforming growth factor β1 signal is crucial for dedifferentiation of cancer cells to cancer stem cells in osteosarcoma. Stem Cells. 2013;31:433-446. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 83] [Cited by in F6Publishing: 85] [Article Influence: 8.5] [Reference Citation Analysis (0)] |
| 52. | Akunuru S, James Zhai Q, Zheng Y. Non-small cell lung cancer stem/progenitor cells are enriched in multiple distinct phenotypic subpopulations and exhibit plasticity. Cell Death Dis. 2012;3:e352. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 90] [Cited by in F6Publishing: 98] [Article Influence: 8.2] [Reference Citation Analysis (0)] |
| 53. | Ning X, Du Y, Ben Q, Huang L, He X, Gong Y, Gao J, Wu H, Man X, Jin J, Xu M, Li Z. Bulk pancreatic cancer cells can convert into cancer stem cells(CSCs) in vitro and 2 compounds can target these CSCs. Cell Cycle. 2016;15:403-412. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 32] [Cited by in F6Publishing: 28] [Article Influence: 3.5] [Reference Citation Analysis (0)] |
| 54. | Kobayashi S, Yamada-Okabe H, Suzuki M, Natori O, Kato A, Matsubara K, Jau Chen Y, Yamazaki M, Funahashi S, Yoshida K, Hashimoto E, Watanabe Y, Mutoh H, Ashihara M, Kato C, Watanabe T, Yoshikubo T, Tamaoki N, Ochiya T, Kuroda M, Levine AJ, Yamazaki T. LGR5-positive colon cancer stem cells interconvert with drug-resistant LGR5-negative cells and are capable of tumor reconstitution. Stem Cells. 2012;30:2631-2644. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 112] [Cited by in F6Publishing: 121] [Article Influence: 11.0] [Reference Citation Analysis (0)] |
| 55. | Chaffer CL, Brueckmann I, Scheel C, Kaestli AJ, Wiggins PA, Rodrigues LO, Brooks M, Reinhardt F, Su Y, Polyak K, Arendt LM, Kuperwasser C, Bierie B, Weinberg RA. Normal and neoplastic nonstem cells can spontaneously convert to a stem-like state. Proc Natl Acad Sci USA. 2011;108:7950-7955. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 842] [Cited by in F6Publishing: 850] [Article Influence: 65.4] [Reference Citation Analysis (0)] |
| 56. | Kaushik V, Yakisich JS, Way LF, Azad N, Iyer AKV. Chemoresistance of cancer floating cells is independent of their ability to form 3D structures: Implications for anticancer drug screening. J Cell Physiol. 2019;234:4445-4453. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 7] [Cited by in F6Publishing: 8] [Article Influence: 1.3] [Reference Citation Analysis (0)] |
| 57. | Yakisich JS, Azad N, Kaushik V, Iyer AKV. Cancer Cell Plasticity: Rapid Reversal of Chemosensitivity and Expression of Stemness Markers in Lung and Breast Cancer Tumorspheres. J Cell Physiol. 2017;232:2280-2286. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 15] [Cited by in F6Publishing: 16] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 58. | Chen X, Wang Y, Feng T, Yi M, Zhang X, Zhou D. The overshoot and phenotypic equilibrium in characterizing cancer dynamics of reversible phenotypic plasticity. J Theor Biol. 2016;390:40-49. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 9] [Cited by in F6Publishing: 14] [Article Influence: 1.6] [Reference Citation Analysis (0)] |
| 59. | Zhou D, Wang Y, Wu B. A multi-phenotypic cancer model with cell plasticity. J Theor Biol. 2014;357:35-45. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 21] [Cited by in F6Publishing: 23] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 60. | Zhou D, Wu D, Li Z, Qian M, Zhang MQ. Population dynamics of cancer cells with cell state conversions. Quant Biol. 2013;1:201-208. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 20] [Cited by in F6Publishing: 20] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 61. | Zheng X, Shen G, Yang X, Liu W. Most C6 cells are cancer stem cells: evidence from clonal and population analyses. Cancer Res. 2007;67:3691-3697. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 163] [Cited by in F6Publishing: 163] [Article Influence: 9.6] [Reference Citation Analysis (0)] |
| 62. | Dirkse A, Golebiewska A, Buder T, Nazarov PV, Muller A, Poovathingal S, Brons NHC, Leite S, Sauvageot N, Sarkisjan D, Seyfrid M, Fritah S, Stieber D, Michelucci A, Hertel F, Herold-Mende C, Azuaje F, Skupin A, Bjerkvig R, Deutsch A, Voss-Böhme A, Niclou SP. Stem cell-associated heterogeneity in Glioblastoma results from intrinsic tumor plasticity shaped by the microenvironment. Nat Commun. 2019;10:1787. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 251] [Cited by in F6Publishing: 299] [Article Influence: 59.8] [Reference Citation Analysis (0)] |
| 63. | Vainstein V, Kirnasovsky OU, Kogan Y, Agur Z. Strategies for cancer stem cell elimination: insights from mathematical modeling. J Theor Biol. 2012;298:32-41. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 39] [Article Influence: 3.3] [Reference Citation Analysis (0)] |
| 64. | Dilão R. Chemotherapy in heterogeneous cultures of cancer cells with interconversion. Phys Biol. 2014;12:016002. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 3] [Cited by in F6Publishing: 3] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 65. | Lefrère F, Delmer A, Levy V, Delarue R, Varet B, Hermine O. Sequential chemotherapy regimens followed by high-dose therapy with stem cell transplantation in mantle cell lymphoma: an update of a prospective study. Haematologica. 2004;89:1275-1276. [PubMed] [Cited in This Article: ] |
| 66. | Tao S, Song L, Deng Y, Chen Y, Gan Y, Li Y, Ding Y, Zhang Z, Ding B, He Z, Wang C, Yu L. Successful treatment of two relapsed patients with t(11;19)(q23;p13) acute myeloid leukemia by CLAE chemotherapy sequential with allogeneic hematopoietic stem cell transplantation: Case reports. Oncol Lett. 2021;21:178. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 1] [Cited by in F6Publishing: 1] [Article Influence: 0.3] [Reference Citation Analysis (0)] |
| 67. | Liu Z, Wang Y, Shan F, Ying X, Zhang Y, Li S, Jia Y, Li Z, Ji J. 5-Fu-Based Doublet Regimen in Patients Receiving Perioperative or Postoperative Chemotherapy for Locally Advanced Gastric Cancer: When to Start and How Long Should the Regimen Last? Cancer Manag Res. 2021;13:147-161. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 5] [Article Influence: 1.7] [Reference Citation Analysis (0)] |
| 68. | Kunzmann V, Siveke JT, Algül H, Goekkurt E, Siegler G, Martens U, Waldschmidt D, Pelzer U, Fuchs M, Kullmann F, Boeck S, Ettrich TJ, Held S, Keller R, Klein I, Germer CT, Stein H, Friess H, Bahra M, Jakobs R, Hartlapp I, Heinemann V; German Pancreatic Cancer Working Group (AIO-PAK) and NEOLAP investigators. Nab-paclitaxel plus gemcitabine vs nab-paclitaxel plus gemcitabine followed by FOLFIRINOX induction chemotherapy in locally advanced pancreatic cancer (NEOLAP-AIO-PAK-0113): a multicentre, randomised, phase 2 trial. Lancet Gastroenterol Hepatol. 2021;6:128-138. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 37] [Cited by in F6Publishing: 69] [Article Influence: 17.3] [Reference Citation Analysis (0)] |
| 69. | Nakamura A, Yamaguchi T, Ito R, Kawakami K. [Successful disease control of plasma cell leukemia by the treatment comprising proteasome inhibitors, followed by daratumumab, lenalidomide, and dexamethasone therapy]. Rinsho Ketsueki. 2020;61:1600-1604. [PubMed] [DOI] [Cited in This Article: ] [Cited by in F6Publishing: 1] [Reference Citation Analysis (0)] |
| 70. | Yu R, Bai H, Gao B, Li T, He X, Zhang P, Wang J. Rare case of apatinib acquired resistance induced by point mutation of WRN p.V697F through activation of the PI3K/AKT apoptosis-inhibiting pathway. Thorac Cancer. 2021;12:128-132. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 7] [Article Influence: 2.3] [Reference Citation Analysis (0)] |
| 71. | Lim B, Song J, Ibrahim NK, Koenig KB, Chavez-MacGregor M, Ensor JE Jr, Gomez JS, Krishnamurthy S, Caudle AS, Shaitelman SF, Whitman GJ, Valero V. A Randomized Phase II Study of Sequential Eribulin Versus Paclitaxel Followed by FAC/FEC as Neoadjuvant Therapy in Patients with Operable HER2-Negative Breast Cancer. Oncologist. 2021;26:e230-e240. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 2] [Cited by in F6Publishing: 2] [Article Influence: 0.5] [Reference Citation Analysis (0)] |
| 72. | Marra A, Curigliano G. Adjuvant and Neoadjuvant Treatment of Triple-Negative Breast Cancer With Chemotherapy. Cancer J. 2021;27:41-49. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 6] [Cited by in F6Publishing: 18] [Article Influence: 6.0] [Reference Citation Analysis (0)] |
| 73. | Huang Y, Wu H, Li X. Novel sequential treatment with palbociclib enhances the effect of cisplatin in RB-proficient triple-negative breast cancer. Cancer Cell Int. 2020;20:501. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 5] [Cited by in F6Publishing: 7] [Article Influence: 1.8] [Reference Citation Analysis (0)] |
| 74. | Delwar ZM, Avramidis D, Siden Å, Cruz M, Yakisich JS. Depletion of drug-surviving glioma cells by a second phase treatment with low concentration of salinomycin. Drug Ther Stud. 2011;1:e7. [Cited in This Article: ] |
| 75. | Ferruzzi P, Mennillo F, De Rosa A, Giordano C, Rossi M, Benedetti G, Magrini R, Pericot Mohr Gl, Miragliotta V, Magnoni L, Mori E, Thomas R, Tunici P, Bakker A. In vitro and in vivo characterization of a novel Hedgehog signaling antagonist in human glioblastoma cell lines. Int J Cancer. 2012;131:E33-E44. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 35] [Cited by in F6Publishing: 37] [Article Influence: 3.1] [Reference Citation Analysis (0)] |
| 76. | Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646-674. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39812] [Cited by in F6Publishing: 42985] [Article Influence: 3306.5] [Reference Citation Analysis (4)] |
| 77. | Dale Rein I, Solberg Landsverk K, Micci F, Patzke S, Stokke T. Replication-induced DNA damage after PARP inhibition causes G2 delay, and cell line-dependent apoptosis, necrosis and multinucleation. Cell Cycle. 2015;14:3248-3260. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 39] [Cited by in F6Publishing: 39] [Article Influence: 4.9] [Reference Citation Analysis (0)] |
| 78. | Lloyd RL, Wijnhoven PWG, Ramos-Montoya A, Wilson Z, Illuzzi G, Falenta K, Jones GN, James N, Chabbert CD, Stott J, Dean E, Lau A, Young LA. Combined PARP and ATR inhibition potentiates genome instability and cell death in ATM-deficient cancer cells. Oncogene. 2020;39:4869-4883. [PubMed] [DOI] [Cited in This Article: ] [Cited by in Crossref: 116] [Cited by in F6Publishing: 97] [Article Influence: 24.3] [Reference Citation Analysis (0)] |