Abstract
Gene drives are selfish genetic elements that are transmitted to progeny at super-Mendelian (>50%) frequencies. Recently developed CRISPR–Cas9-based gene-drive systems are highly efficient in laboratory settings, offering the potential to reduce the prevalence of vector-borne diseases, crop pests and non-native invasive species. However, concerns have been raised regarding the potential unintended impacts of gene-drive systems. This Review summarizes the phenomenal progress in this field, focusing on optimal design features for full-drive elements (drives with linked Cas9 and guide RNA components) that either suppress target mosquito populations or modify them to prevent pathogen transmission, allelic drives for updating genetic elements, mitigating strategies including trans-complementing split-drives and genetic neutralizing elements, and the adaptation of drive technology to other organisms. These scientific advances, combined with ethical and social considerations, will facilitate the transparent and responsible advancement of these technologies towards field implementation.
Similar content being viewed by others
Introduction
Exploiting genetic systems that link desired traits to chromosomes or genetic elements with a positive transmission bias (that is, >50%) dates back to the potential uses of chromosomal translocations by Serebrovski1, which was further generalized and articulated by Curtis in the 1960s for spreading a desired trait throughout a target population2. These so-called gene-drive systems or selfish genes3 are abundant in nature. Driving elements can bias the transmission of sex chromosomes or autosomes (meiotic drive)4,5,6,7,8,9,10,11 or only themselves, as exemplified by the diverse families of transposable elements12,13 (for example, P-elements in fruitflies14,15,16 or retrotransposons in humans17). Such super-Mendelian genetic entities have been implicated in the evolution of genome architectures in plants and animals17,18,19,20,21.
Gene drives can be broadly divided into two main categories based on how readily they spread through a population. High-threshold drives, such as the reciprocal chromosomal translocations that Curtis considered2, require many individuals (for example, more than the number of native residents) to take over the population (Fig. 1). By contrast, low-threshold drives can be seeded at very low numbers to do so. This Review focuses on the latter low-threshold gene drives in insects as they arguably hold the greatest promise for impacting disease transmission on continental scales and because optimized second-generation drives have been developed over the past 5 years. Various high-threshold systems, with diverse applications on more local scales, are reviewed elsewhere, including second-generation so-called underdominant chromosome translocation systems22 and other strategies such as Medea toxin–antitoxin arrangements23,24,25,26,27,28,29, Cleave and Rescue30,31 or TARE systems32, in which an essential gene is targeted for inactivation and rescued by a recoded transgene inserted elsewhere in the genome, or reproductive symbionts/parasites such as Wolbachia33,34,35.
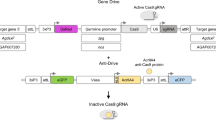
Gene-drive scheme68. The bipartite synthetic CRISPR system (part a). A guide RNA (gRNA; green) binds Cas9 (cyan) directing it to bind and cleave DNA at complementary sites 20 nucleotides in length. The protospacer-adjacent motif (PAM) site (NGG; red) is required for Cas9 binding to genomic targets. In eukaryotic cells, double-stranded breaks are repaired either by the error-prone non-homologous end-joining or by homology-directed repair (HDR), the pathway acting in the germline (part b). Insertion of a cassette encoding Cas9 (cyan) and a gRNA (green) flanked by homology arms (HAs) results in HDR-mediated copying of the cassette from the plasmid into the genomic cut site (part c). The HAs directly flank the gRNA-directed cleavage site. Once inserted into the genome, the Cas9 + gRNA cassette directs cleavage of the homologous chromosome in the germline and is copied into the DNA break by HDR resulting in nearly all progeny (~99%) inheriting the ‘gene-drive’ cassette (part d). Comparison of Mendelian versus gene-drive inheritance patterns. In each case, a few transgenic individuals (blue) are introduced to a large wild-type (WT) population (white) (part e). Predicted logistic growth curve for seeding 1% gene-drive individuals into a WT population (part f). This logistic growth curve is defined by the second-order recursion formula: fn+1 = fn + fn(1 – fn) = 2fn – fn2, where fn is the frequency of the gene drive in the population at generation n. This formula has the closed-form solution \({\rm{f}}({\rm{n}}){\rm{=}}1-(1-{{\rm{c}}}_{0}{)}^{({2}^{{\rm{n}}})}\), where c0 is the seeding frequency of the gene drive68. Such optimal drives should reach nearly full introduction by ~10 generations when seeded at a ratio of 1:100. Foundational modelling by Curtis2 for drive of a translocation (T), which is fertile as a homozygote, as is the WT (+) allele, but gives rise to sterile heterozygotes T/+ (part g). This seminal example of a high-threshold drive reveals that, if the T allele is present at >50% prevalence, it takes over the population but, when present at <50%, it disappears over time (solid lines). If the T allele carries a heterozygous fitness cost (dotted lines) the drive must be seeded at a higher percentage to take over. Modelling of a homing endonuclease gene (HEG) gene drive inserted into an essential gene and expressed in a strictly germline-specific fashion63 (part h). Assuming an infinite population and no cleavage-resistant alleles, this drive should attain an equilibrium prevalence determined by its allelic conversion probability (e). For e = 0.9, the drive equilibrates at 90%, at which point the greater reproductive fitness of the WT allele balances the drive potential of the HEG. Parts a–d are adapted with permission from ref.68, Wiley. Part g is adapted from ref.2, Springer Nature Limited. Part h is adapted with permission of The Royal Society, from Proc. Biol. Sci. Burt, A. 270, 1518 (2003); permission conveyed through Copyright Clearance Center, Inc. (ref.63).
Homing endonuclease genes (HEGs)36,37 are an example of low-threshold selfish genetic elements that are found in a variety of microorganisms. These elements encode highly sequence-specific endonucleases that cut a naive homologous chromosome at the site where they are inserted into the genome and are copied into the DNA breaks they create by homology-directed repair (HDR) pathways. HEGs provided the first practical tools for building and testing synthetic gene-drive systems in strains of Drosophila38,39 or Anopheline mosquitoes40 engineered to carry a HEG recognition site.
A limitation of HEGs is that modifying them to cut at specific desired sites in the genome is exceedingly difficult. This problem was bypassed by programmable nucleases, such as zinc-finger nucleases (ZFNs) and transcription activator-like effector nucleases (TALENs), that initiated the era of targeted genome editing41. Subsequently, the discovery of CRISPR–Cas9 bacterial defence systems42 and the development of a simplified dual-component system by Doudna’s group, consisting of the Cas9 (CRISPR-associated protein) endonuclease and a readily programmable guide RNA (gRNA) that binds Cas9 and directs DNA cleavage to desired sites43 (Fig. 1a), has revolutionized nearly all fields of biology44. The bipartite nature and flexible programmability of CRISPR led to the rapid development of a variety of gene-drive systems (Fig. 1b–f) in insects45,46,47,48,49, mammals50, yeast51 and bacteria52, several of which are discussed in this Review.
I begin by discussing full gene-drive systems that carry linked transgenes expressing Cas9 in the germline and a gRNA-directing DNA cleavage at the site where the gene-drive cassette is inserted into the genome45,53. These drive elements can be used to reduce mosquito populations (suppression) or to render them incapable of transmitting pathogens (modification; for example, by including cargo genes encoding anti-malarial effectors46). For modification systems, incorporating functional recoded versions of genes into drives can greatly improve their performance54,55,56,57.
Next, I examine split-drive systems in which the Cas9 and gRNA transgenes are carried by two different elements. Such split systems can be used for various purposes, including deleting and replacing genome segments58, updating gene drives (for example, with new anti-pathogen effectors), reconstituting a full-drive system from separate trans-complementing drive components59, driving transmission of preferred allelic variants at other loci60 (for example, insecticide susceptibility or pathogen refractory host alleles), inactivating host factors required for pathogen transmission61, neutralizing drives (that either excise the drive element or mutate the Cas9 transgene it carries)62, and in promising ‘active genetics’ applications being developed in other organisms50,52. Finally, I consider the wider implications and future perspectives for genetic systems endowed with the inherent capacity to spread throughout natural populations.
Suppression versus modification strategies
There are two primary strategies for deploying low-threshold gene-drive systems to reduce the disease impacts of insect-borne pathogens. The first, often referred to as ‘population suppression’, is the genetic equivalent of insecticides. The idea of suppression drives is to force deleterious traits into a population, leading those populations to crash or be much diminished. If the mosquito is eliminated, so too will all the diseases it can transmit (see Box 1 for the relationship between entomological effects and disease elimination). The second approach is to modify the insect vector to prevent it from transmitting the pathogen one wishes to eliminate. This immunizing approach, often referred to as ‘population modification’ or replacement, leaves the insect in place in the environment but blocks disease transmission.
Mathematical modelling of suppression drives with differing fitness costs by Burt63 (Fig. 1h) predicted that they should reach an equilibrium level in populations of an infinite size determined by the copying efficiency of the drive element. Generational delays in incurring fitness costs improved the suppressive performance of drives (for example, grandchildless > sterile > lethal phenotypes). The main virtue of suppression drives is that, if successful, they eliminate or greatly reduce the transmission of all diseases vectored by a given insect. For example, suppression of Anopheline mosquitoes would reduce malaria caused by all malarial parasites, most notably the main pathogens of concern Plasmodium falciparum and Plasmodium vivax. Likewise, suppression of Aedes mosquitoes, such as Aedes aegypti, would greatly reduce the transmission of all arboviruses vectored by this species, including those causing dengue fever, yellow fever, chikungunya and Zika. The primary challenge of this approach, however, is the possible failure of the drive to achieve its goal of suppression owing to it generating (or the pre-existing presence of) functional cleavage-resistant alleles in the population that cannot be converted by the gene-drive. Such functional drive-resistant alleles would be positively selected, leading to the disappearance of the suppression drive and rebound of drive-resistant vector-competent mosquitoes. Additionally, local elimination of a mosquito species (although modelling suggests this is very unlikely on a global scale64,65; Fig. 2) might result in other species filling in the empty niche66,67, which could have unintended ecological consequences.
a | Genetic map of one of the first CRISPR-based suppression drives inserted into the female-sterile nudel locus in Anopheles gambiae. b | Multigenerational cage studies with the nudel-drive resulted in an initial increase followed by a progressive loss of the gene-drive element without crashing the target population. This drive trajectory was attributed to the generation and selection of functional cleavage-resistant alleles of the target gene that took over owing to them being female fertile. c | Genetic map of the third-generation doublesex (dsx)-shredder drive, which combines both drive at a highly conserved target site in the dsx locus48 with an optimized X-shredder84. d | Comparative modelling of the dual dsx-shredder to the parent dsx-drive. The dual drive (d1) reduces female numbers to a much greater extent than the dsx-drive (d2) and does so more rapidly with less imposed load. e–g | Spatial modelling of a driving-Y system, wherein 500 mosquitoes are released weekly in a grid-like pattern of 15 release sites in the Garki district of Nigeria70. In this particular simulation, the drive spreads successfully throughout the target area and eliminates the mosquito population locally. Green represents a completely wild-type population, whereas darkening shades of purple represent increasing fractions of the drive allele. The outcomes of these simulations depend on various factors, including seasonality and mosquito mobility, which influence the likelihood of wild-type mosquitoes recolonizing the treated region. h,i | Spatial and temporal modelling of a driving-Y released over a large geographical area centred on Burkina Faso64 (part h). In this scenario, 10 transgenic male mosquitoes are released at 1% of all sites (42,360 sites total) each year (sites are chosen randomly each year). Seasonal variations in mosquito numbers (transgenic and wild type) in locals, labelled i1–i6 on the maps, simulate population structures after 4 (left) or 8 (right) years of release. Similar spatial and temporal modelling results were obtained for the release of the CRISPR-based dsx-drive and dsx-shredder drive65. DSB, double-stranded break; gRNA, guide RNA; spCas9, Streptococcus pyogenes Cas9. Parts a and b are reprinted from ref.71, CC BY 4.0 (https://creativecommons.org/licenses/by/4.0/). Parts c and d are reprinted from ref.84, CC BY 4.0 (https://creativecommons.org/licenses/by/4.0/). Parts e–g are reprinted with permission from ref.70, PNAS. Parts h and i are reprinted from ref.64, CC BY 4.0 (https://creativecommons.org/licenses/by/4.0/).
The alternative strategy of population modification, in which drives are designed to carry minimal, if any, fitness costs, are predicted to follow a simple logistic growth trajectory when released into wild populations (Fig. 1e,f). If the drive copies with high efficiency in both sexes (see below for examples of drives approximating this ideal), then, when seeded at ratios as low as 1:10 or perhaps even 1:100, they should completely replace the wild-type allele in 5–10 generations68 (Fig. 1f). The chief advantage of modification drives is that they are predicted to remain stable in the population for a sufficiently long period (2–5 years) to achieve and maintain local elimination of the pathogen (for example, malarial parasite; Box 1), allowing public health officials to “consolidate their gains” as Macias and James, the latter a leader in the development of modification strategies, often point out69. One could then move forwards to rid the disease from neighbouring or more distant regions, to achieve eventual continent-scale elimination of the parasite. This is a salient theoretical advantage of modification over suppression, because suppression, like insecticides, may need to be applied repeatedly in the same location64,65,70, creating escalating costs and logistical barriers as the effort expands. Additionally, modification drives should leave no empty ecological niches66,67 and, therefore, potentially impose less environmental impact than suppression drives. The major limitations of the modification strategy are that anti-pathogen cassettes need to be built for each pathogen (for example, each species of malarial parasite or each target virus) and redundant effector systems are required to avoid the rapid selection for resistance to the anti-pathogen factors (Supplementary Table 1). In some geographical regions, the former consideration could require constructing several transgenic lines in multiple species. Updating genetic elements (described below) can help address the latter concern regarding evolution of parasite resistance.
Suppression drives
First-generation suppression drives developed in the Crisanti laboratory47 (Fig. 2a) performed comparably to the proof-of-principle modification drives46. However, suppression drives, by their intended design, suffer a greater impact from imperfect copying when transmitted through females71. The problem stems from non-copying events in which the target site is cleaved and, instead of the break being repaired by HDR-mediated directional gene conversion72,73,74,75 (resulting in copying of the drive element), the site is mutated by the competing non-homologous end-joining (NHEJ) pathway to generate drive-resistant insertion/deletion (indel) alleles45,46,47,49,60,76,77. Most of such NHEJ-induced indel alleles are non-functional (for example, out-of-frame or deleterious to protein function) and contribute passively to suppression of the target population71. However, a fraction of in-frame indels can retain target gene function and such alleles will rapidly take over the population owing to the strong positive selective advantage associated with fertility (Fig. 2b).
The doublesex-drive
The Crisanti group found a clever solution to the drive-resistance problem by identifying a highly conserved gRNA target site in the doublesex (dsx) gene, which is required for proper female development and, hence, fertility48. This target site was conserved 100% across more than 1,000 sequenced genomes of the target species (Anopheles gambiae)78 and was located at an intron–exon junction. The authors reasoned that this target sequence could not be altered in any fashion without loss of an essential differential splicing product required to generate the female-specific form of this sex-determination protein. Additional improvements, including the identification of a more germline-specific promoter (from the zero population growth (zpg) gene) to express the Cas9 enzyme selectively in cells where HDR prevails, reduced the frequency of generating NHEJ alleles. This second-generation suppression drive produced fewer NHEJ alleles and no functional drive-resistant alleles were identified in cage trials, indicating that the frequency of such alleles is exceedingly low, possibly zero. As no functional drive-resistant alleles arose, the dsx-drive consistently took over cages with wild-type mosquitoes and collapsed the populations as intended.
X-shredder drives
Another potential suppression tactic is to destroy (or shred) X chromosomes in males so that only the Y chromosome is transmitted79. A key aim for this approach is to destroy79 or mutate80 the X chromosome prior to fertilization of the egg to avoid reducing total progeny output. Note that one could also express the X-shredder from an autosome to generate daughterless fathers, however, as this sterilizing trait would be inherited in only a Mendelian fashion, such elements would rapidly decrease in frequency in the population in contrast to exhibiting drive when linked to the Y chromosome. Proof-of-principle X-shredders using either HEG or CRISPR systems targeting repeated X-linked ribosomal RNA genes showed promise81,82; however, it proved difficult to express the nucleases from the Y chromosome, most likely owing to the phenomenon of meiotic sex chromosome inactivation83. Another general concern for these and other suppression drives is to minimize any dominant fitness costs associated with being heterozygous for the drive element.
A composite dsx-shredder drive
Although the dsx-drive very effectively suppressed laboratory cage populations without generating functional drive-resistant alleles48, there was the lingering concern that such alleles might arise very rarely in large natural populations. One way to reduce this possibility is to impose female sterility by two independent mechanisms. The Crisanti group created such a dual sterilization drive by mounting a HEG X-shredder under the control of a male-specific promoter (beta2-tubulin) on the dsx-drive84 (Fig. 2c). In the process, they also optimized regulatory sequences to minimize the heterozygous fitness costs of the drive element. Both mathematical modelling and cage experiments confirmed that the composite dsx-shredder drove more quickly than the parent dsx-drive (Fig. 2d).
Modelling on geographical scales
Modelling efficient suppression systems, such as the dsx-drive and dsx-shredders, predicts the observed rapid collapse of populations observed in cage studies involving limited population sizes48,84. Recall that original models devised by Burt63, assuming much larger populations, concluded that such elements would achieve a balanced equilibrium based on drive efficiency versus fitness cost associated with the drive relative to wild-type alleles (Fig. 1h). More recent two-dimensional models accounting for realistic population densities of mosquitoes on large geographical scales reveal again that suppression drives should not fully extinguish themselves even long after they have spread throughout a region64,65,84 (Fig. 2h). These models predict periodic regional fluctuations of mosquito populations (Fig. 2i), wherein both driving Y64 or dsx (± X-shredding) suppression65 drives achieve complete population elimination in local areas (as they do in confined population cages48,84 or in circumscribed territories following grid-like patterns of release70; Fig. 2e–g). Because mosquitoes homozygous for the drive are not able to repopulate those regions (they are female sterile), only wild-type mosquitoes flying in from adjacent regions can do so64,65. Those wild-type mosquitoes can then breed until they achieve sufficient densities to sustain re-introduction of the suppression drive, which also never goes extinct because wild-type populations keep resurging in oscillating patterns of territories (Fig. 2i). The final outcome is that the average number of mosquitoes is greatly reduced (by ~95% with ideal drive performance) but the fluctuating equilibrium between drive and wild-type mosquito populations persists indefinitely, always ebbing and flowing, particularly in regions with dense mosquito and human populations64,65 (Fig. 2i). The practical question is: are these levels of suppression sufficient to bring the basic reproductive number (R0) of parasite infection below 1, the epidemiological criterion for extinguishing an epidemic (Box 1)?
Modification drives
In contrast to suppression drives, the impediment to drive success imposed by functional alleles resistant to Cas9 cleavage is much less of a concern for modification drives, which are designed not to impose any significant fitness costs70. Consistent with the expectation that rare functional drive-resistant alleles should have little, if any, advantage over a fitness-neutral drive allele, such variants have not been observed to expand in small population cages54,57,85, in contrast to the strong positive selection imposed on this class of alleles by suppression drives47,86. These comparative considerations notwithstanding, it still makes good engineering sense for modification drives to achieve maximal introduction into a target population. Such deep population penetration is important for two reasons. First, protective effectors, such as transmission-blocking single-chain antibodies (scFvs) that bind to and neutralize malarial parasites (Supplementary Table 1) need to be present in a sufficiently large fraction of the population to attenuate disease transmission such that R0 <1 (Box 1). Second, for some effectors, it may be beneficial or necessary for them to be present in two copies (that is, homozygous) to achieve sufficient levels of effector dosage to have the desired effect87,88. In such cases, one needs correspondingly high levels of drive introduction to achieve the desired goal otherwise a considerable fraction of the population may remain heterozygous for the effector cassette and cleavage-resistant NHEJ alleles (see below regarding recoded drives inserted into essential genes to avoid such outcomes). As in the case of suppression drives, one would expect comparable epidemiological impacts in mosquito populations if effectors blocking 100% of parasite transmission (as they do in laboratory settings89) were carried by a sufficient fraction of the population (for example, an 80% reduction in mosquito numbers ≈ 80% prevalence of fully effective anti-parasite effectors; Box 1). However, if parasite transmission were not entirely attenuated, the fraction of modified individuals might need to be higher to achieve similar reductions in R0.
Three potential types of fitness-neutral modification drive
Whereas the impact of functional drive-resistant alleles is very different for suppression drives (potentially debilitating) versus modification drives (an optimization problem), the molecular mechanisms underlying the generation of such alleles is the same. The problem is with female-mediated drive transmission, which stems from maternal deposition of Cas9–gRNA complexes into the egg45,46,47,49,60,76,77. These perduring ribonucleoprotein complexes can then act on the paternal allele following fertilization but preceding allocation of the germline. If a paternal allele successfully avoids NHEJ mutagenesis and survives intact to enter the germline compartment, the drive process is nearly 100% efficient because germline cells repair DNA breaks almost exclusively via the HDR pathway (which is why sexual reproduction is not mutagenic)72,73,74,75. Similar double-strand breaks generated by host factors during meiotic recombination are repaired by HDR to resolve crossover events90.
There are three general solutions to drive attenuation in females. One solution is to restrict drive to males, thereby not generating any DNA cleavage in the female germline that could damage paternal alleles. Although simple in principle, this strategy requires the identification of suitable male-specific regulatory elements to control Cas9 expression, which remain to be characterized. The second solution is to limit the expression of the Cas9 nuclease to the period of germline development (for example, meiosis) when HDR takes place and then shut it off abruptly prior to the stages at which it would accumulate in the egg (for example, in nurse cells)85,91. Such restricted Cas9 expression should, in principle, result in a drive that is transmitted ~100% of the time through both male and female lineages (see A clean drive below). The third solution is to eliminate individuals carrying non-drive NHEJ events, that is, to kill or sterilize all the mistakes. This latter strategy, which can be highly effective and offers a general solution to the drive-resistance problem, depends on a phenomenon referred to as lethal or sterile mosaicism (see below and Box 2).
A clean drive
Male and female mosquitoes carrying a gene drive inherited from their fathers can transmit that element to ~99% of their progeny. Thus, in principle, it should also be possible to achieve similar transmission if the drive were inherited from their mothers so long as significant levels of Cas9–gRNA complexes were not deposited into the eggs85,91. Although several of the mosquito drives described above use the germline-specific nanos or zpg promoters to express Cas9 and minimize this problem91, they all still generate some appreciable rate of NHEJ alleles (for the dsx-drive, none of those alleles was functional). One example of a drive that greatly minimizes the generation of such drive resistance is inserted into the cardinal (cd) locus of An. gambiae in which the Cas9 transgene is expressed under the control of nanos regulatory sequences85. The cd-drive is remarkable in that it generates virtually no NHEJ alleles of any kind and, when homozygous, has little if any fitness effects (other than a transient loss of eye pigmentation during larval stages). It is not yet clear whether this highly efficient performance can be achieved at other loci or in other species using the same (or orthologous) Cas9 control sequences or whether there is something special about the cd locus or the gRNA it uses to copy. Those important issues notwithstanding, the cd-targeted drive provides unequivocal evidence that such high-performing drives inserted into fitness-neutral locations in the genome are possible to develop and perform comparably even when loaded with substantial cargo such as dual anti-malarial scFvs (A. A. James, personal communication) (Supplementary Table 1). In population cages, the cd-drive system can attain full introduction when seeded with 5% drive alleles in only seven generations, following a predicted logistical growth trajectory85. The cd-drive is likely to be one of the systems advanced for consideration in the next phase of testing in open field trials (see below).
Recoded drives inserted into essential genes
Although the encouraging example of the cd-drive demonstrates that it is possible to copy with high efficiency in both sexes, it is not clear how general such strategies will prove to be or whether they can be readily transportable between species in which regulatory elements may have evolved extensively (for example, beyond ~50 million years of species divergence). It is also challenging to identify highly conserved gRNA target sites in fitness-neutral loci (for example, cd) or in non-coding sequences that display high degrees of sequence variation78,92,93,94,95. However, difficulties posed by the latter issue may have been overestimated as a recent study has found that such highly conserved sequences can be readily identified in nearly every locus if one searches for them appropriately96.
A more general solution to the maternal deposition problem is to kill or sterilize all non-copying events in which non-functional NHEJ alleles are generated (Box 2). The idea is to insert a gene drive into a locus that is essential for the survival or fertility of the organism and to have the drive element also provide a functional copy of that gene consisting of in-frame cDNA sequences that have been recoded to avoid spurious recombination with the endogenous gene. Such recoded gene-drive elements confer viability and fertility in a homozygous state30,31,32,53,54,55,56,57,61, whereas deleterious recessive NHEJ alleles assume dominant lethal or sterile phenotypes during the drive process and are culled (Box 2).
An example of a recoded lethal/sterile mosaic drive has recently been developed in the kynurenine hydroxylase (kh) locus in Anopheles stephensi54 (Fig. 3). This khRec-drive (Fig. 3d) is a second-generation version of the prototype non-recoded kh-drive (Fig. 3a), which provided the first proof of principle for CRISPR-based gene drives in mosquitoes46. Individuals homozygous for either the original kh-drive or loss-of-function NHEJ alleles have white eyes and such females are effectively sterile (most die after a blood meal and those that survive produce few progeny86). Whereas efficient transmission via males sustained a significant level of drive introduction for the non-recoded element in cage experiments86, appreciable generation of NHEJ alleles in females resulted in the drive either collapsing populations (like the suppression drives described above) or being outcompeted by functional NHEJ alleles (Fig. 3b). However, the khRec-drive fuses a functional recoded kh cDNA in-frame at the insertion site of the drive element (Fig. 3d). As this insertion site is in an enzymatically critical region of the gene, nearly all NHEJ alleles are loss-of-function. When this drive was seeded, even at low frequencies (~5% allelic prevalence), into population cages it rapidly drove to >95% introduction and, as it did so, generated a burst of white-eyed sterile females in which otherwise recessive NHEJ alleles were converted into dominance by lethal/sterile mosaicism (Box 2). NHEJ alleles transmitted to males by drive-carrying mothers were eventually eliminated by the slower traditional process wherein females homozygous for recessive kh– NHEJ alleles suffer the fitness burdens described above. The combined action of these two selective processes led to rapid sustained elimination of non-functional NHEJ alleles and efficient khRec-drive (Fig. 3c).
a | Genetic map of the first CRISPR-based modification gene drive (kh-drive46) in mosquitoes. The construct contains the anti-malarial short-chain variable fragment (scFv) genes m2A10 and m1C3 and was inserted into the kynurenine hydroxylase (kh) locus in Anopheles stephensi. Mosquitoes homozygous for this gene-disrupting kh-drive element or for loss-of-function non-homologous end-joining (NHEJ) alleles have white eyes and are female sterile. b | Multigenerational cage studies with the kh-drive86 reveal a similar outcome to those with the suppression drive shown in Fig. 2b. At high seeding ratios (1:1) the drive increases and, in two out three replicates, crashes the cage. In the other 1:1 replicate as well as in all three cages seeded at only a 1:9 ratio, the drive increases for a few generations and then progressively disappears owing to accumulation of, and selection for, functional cleavage-resistant NHEJ alleles. c | In multigenerational studies, the kh-recoded drive54 successfully attains high levels in all cage replicates seeded at either 1:1 or 1:9 seeding ratios. At the steepest portion of the curve, bursts of white-eyed non-fluorescent (that is, NHEJ) alleles are generated by the process of lethal/sterile mosaicism, as indicated by the asterisks and white eye photos. d | Genetic map of kh-recoded drive, which is inserted into the exact same site as the original non-recoded kh-drive shown in part a. The recoded drive was generated by a cassette-swapping protocol that replaced the DsRed marker with GFP. cDNA, complementary DNA; gRNA, guide RNA; HDR, homology-directed repair. Part a is adapted with permission from ref.46, PNAS. Part b is reprinted from ref.86, CC BY 4.0 (https://creativecommons.org/licenses/by/4.0/). Parts c and d are adapted from 54), CC BY 4.0 (https://creativecommons.org/licenses/by/4.0).
The great advantage of the recoded lethal/sterile mosaic mechanism is that it should be readily generalizable to a broad variety of essential genes55,56,57 in diverse species, thereby eliminating the need to identify narrowly defined conditions required to design an optimal drive. However, an important requirement for this strategy is that the drive is inserted at a site in the target gene that is critical for its function (Box 2).
Alternative drive systems
In addition to providing the exquisite sequence specificity needed for inserting gene drives into particular genomic locations, CRISPR offers another substantial advantage to this field, namely that the active nuclease is comprised of separate Cas9 and gRNA components. One can exploit this bipartite nature to generate a variety of flexible systems such as gRNA-only updating drives, which carry gRNAs ± cargo but no source of Cas9. These split-drive systems include CHACRs (constructs hitchhiking on the autocatalytic chain reaction) used in combination with autonomous full drives58,68, spilt-recoded drives powered by separate static Cas9 sources inherited in a Mendelian fashion located elsewhere in the genome55,56,57,61, trans-complementing split-drives59 and allelic drives60 that bias the inheritance of preferred allelic variants at separate genomic locations. Such split systems offer flexibility and potentially less invasive forms of drive that may be suitable to more localized applications requiring less stringent laboratory confinement97.
Drive-updating CHACR elements
Once a gene-drive system has attained full introduction into a population (nearly every organism carrying at least one copy of the drive system86), it may become necessary to update its functions. For example, in the case of modification drives, if the targeted malarial parasite becomes resistant to the effectors carried by a drive, it may be desirable to introduce new combinations of transmission-blocking effectors targeting additional parasite epitopes and infection stages87,89. Other new functionalities could also include gRNAs that bias the inheritance of desired allelic variants such as those conferring susceptibility to existing insecticides in regions where resistance to those chemicals poses a substantial vector-control challenge (see the Allelic drives section below).
A simple updating element is a CHACR, which carries a gRNA targeting its insertion into the genome, a dominant marker and the desired cargo58,68. If released into a gene drive-bearing population, CHACRs should follow the same logistical growth dynamics as a gene drive does in a naive population. Like full drives, CHACRs can be inserted into fitness-neutral sites or, alternatively, into essential genes, in which case, the CHACR would also carry recoded cDNAs to restore gene function. For example, the FREP1 gene of mosquitoes is required by malarial parasites to infect the midgut tissue and mutations in this gene greatly reduce parasite transmission98,99,100,101. This gene also plays an important role in the host as FREP1–/– mutants suffer severe fitness costs101. However, an allelic variant of FREP1 in An. gambiae sustains wild-type gene function but abrogates binding to the parasite98,99,100. One could therefore insert a CHACR into the FREP1 locus (or similarly for any other gene encoding an essential host factor102) carrying a recoded parasite-resistant allele and possibly additional anti-malarial effector cassettes to assault the parasite with a cocktail of effectors87. This type of combinatorial strategy is likely to be essential in preventing parasites from rapidly evolving resistance to individual or even pairs of effectors. CHACRs can also carry additional gRNAs to target other sites in the genome for mutagenesis61 (for example, the e-CHACR drive-neutralizing system described below62), delete and replace segments of the genome (using two gRNAs)58, insert cargo plus a gRNA into an intron of a target gene followed by a 2A ribosome-skipping sequence to retain activity of the endogenous locus (integral gene drives)103, or bias inheritance of beneficial allelic variants60 (see the Allelic drives section below).
Trans-complementing drives
One potential concern regarding full gene drives is that individuals carrying these elements might somehow escape prior to an approved release event97. Additionally, testing different effector molecules for function requires greater regulatory oversight and more stringent confinement measures using full versus split systems103. One way to get the best of both worlds, benefitting from efficient drive and low risk of premature drive release, is to use a trans-complementing design in which the Cas9-bearing and gRNA-bearing elements are inserted at two different locations in the genome59. The gRNA element encodes two gRNAs (± cargo): one cutting at its own site of genomic integration to copy in the presence of a Cas9 source acting in trans and the other gRNA cutting at the site where the Cas9 transgene is inserted (Fig. 4a). On their own, either element is inherited in a Mendelian fashion but, when combined, the two gRNAs carried by the gRNA element result in the Cas9-dependent copying of both elements. Trans-complementing drives and the integral gene drive system mentioned above also offer advantages for field testing under non-driving conditions103 as, in both of these systems, components carrying the effector cassettes can be maintained as simple Mendelian strains.
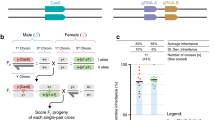
a | A trans-complementing drive system59 comprises two constructs, each of which alone is inherited in a standard Mendelian fashion. The first element carries a Cas9 transgene expressed in the germline and the second carries two guide RNAs (gRNAs), one to copy itself and the other to cut at the genomic site of Cas9 insertion (to copy that element). When combined, the gRNAs carried by the second element drive super-Mendelian inheritance of both elements. b,c | Schemes for allelic-drive60. A drive element (in this case, a split drive) carries one gRNA to copy itself (yellow) and a second (blue or purple) to cleave a non-preferred allele (for example, the DDT-resistant ENa sodium ion-channel 1014F allele) but not the preferred wild-type (ENa 1014L) allele (location indicated by thick blue line). Similarly, the parasite-refractory FREP1 442Q allele could be driven by a gRNA selectively cutting the parasite-sustaining 442L allele. Copy-cutting is the simplest form of allelic-drive, in which the gRNA directly cuts the non-preferred allele. Copy-grafting is a more general form of allelic-drive in which the preferred allele (location indicated by thick blue line) is associated with a nearby (<25 bp) cleavage-resistant site (engineered or naturally occurring; indicated by the padlock) that cannot be targeted by a gRNA that cuts an otherwise highly conserved sequence present both in the non-preferred and wild-type genetic backgrounds. In combination with Cas9, the gRNA will replace the target sequence with a preferred cleavage-resistant site. d | Scheme illustrating the Cas9-neutralizing action of an e-CHACR (erasing construct hitchhiking on the autocatalytic chain reaction) on a Cas9-bearing gene-drive element (MCR-GFP) inserted at the yellow locus of Drosophila melanogaster. The MCR-GFP element carries a gRNA (green) to copy itself62. An e-CHACR, located at the white, ebony or knirps loci, expresses a gRNA for copying itself (blue) and for mutagenizing Cas9 (purple). Homologous sequences adjacent to the MCR-GFP and e-CHACR elements are indicated by brown versus blue lightly shaded boxes, respectively. e | Graphs of multigeneration experiments in which an e-CHACR (red curves) inserted at the white locus was combined 1:1 with the MCR-GFP drive (green curves). Orange curves denote individuals carrying both the e-CHACR and MCR-GFP elements and yellow curves denote those individuals with active Cas9 revealed by mosaic eye phenotypes. Faintly coloured curves indicate simulations based on mathematical modelling. f | Scheme illustrating an ERACR (element reversing the autocatalytic chain reaction) carrying a recoded yellow gene deleting and replacing the MCR-GFP drive element inserted at the yellow locus62. Asterisks indicate gRNA cut sites ablated by insertion of a gene cassette. g | Graphs of multigeneration experiments in which an ERACR (± recoded yellow sequences) was combined with the MCR-GFP drive at a 1:3 ratio. In all cage replicates, the yellow– ERACR-min (red curves, left panel) drove to near completion while greatly reducing the prevalence of the gene-drive (green curves) after 10 generations. The yellow+ ERACR-2 (red curves, right panel) drove to full introduction and completely eliminated the MCR-GFP element by generation 7, displaying surprisingly little variation between cage replicates. The faintly coloured curves indicate simulations based on mathematical modelling. HDR, homology-directed repair. Part a is adapted from ref.59, CC BY 4.0 (https://creativecommons.org/licenses/by/4.0/). Parts b and c are adapted from ref.60, CC BY 4.0 (https://creativecommons.org/licenses/by/4.0/). Parts d–g are adapted with permission from ref.62, Elsevier.
Allelic drives
Many naturally occurring desired traits result from allelic variants of essential genes. For example, resistance to insecticides104 is often conferred by allelic variants of genes encoding proteins targeted by these chemicals such as ion channels105,106, neurotransmitter receptors107,108 or enzymes modifying neurotransmitter activity109. The wild-type alleles of these genes thus confer susceptibility to standard vector-control measures. Additionally, as mentioned above, an allelic variant of the endogenous FREP1 protein, which is expressed in the mosquito gut, reduces malarial parasite transmission presumably by binding less well to parasites to aid them in transiting the midgut epithelium into the body cavity99,100,101.
Gene drives or updating CHACR elements can carry additional gRNAs that bias the inheritance of favourable allelic variants (allelic drive) by one of two modes: copy-cutting (Fig. 4b) or copy-grafting60 (Fig. 4c). For copy-cutting, a gRNA selectively targets cleavage of the non-preferred allele (for example, one conferring insecticide resistance). In individuals carrying an allelic drive (plus Cas9) and the preferred cleavage-resistant allele of the targeted locus in trans to the cleavable undesired allele, the undesired allele is cut and then repaired using the preferred allele as a correction template. The result is super-Mendelian inheritance of the preferred allele. An important requirement for allelic drive is that non-functional NHEJ alleles are dominantly eliminated by lethal mosaicism, which in turn restricts this type of scheme to driving preferred alleles of essential genes60. Otherwise, cleavage-resistant NHEJ alleles themselves could be driven in a runaway fashion.
A limitation of the copy-cutting strategy is that one needs to be able to design a gRNA that distinguishes between preferred and undesired allelic variants. For standardly used Streptococcus pyogenes Cas9 (spCas9), with its NGG protospacer-adjacent motif (PAM) target sequence requirements, such gRNAs can only be designed approximately half the time. However, Cas9 variants have been developed recently that have more relaxed PAM site restrictions110,111, which could expand the range of targets accessible to copy-cutting. A caveat in using relaxed PAM nucleases is that they would still need to retain sufficient target specificity to avoid cutting the PAM-less gRNA-encoding sequences carried by the drive.
Copy-grafting is a more generally applicable form of allelic drive in which the preferred allele is associated with a neighbouring non-cleavable sequence (~25 bp away)60 (Fig. 4c). If the preferred allele lies within this narrow window of high-frequency HDR-mediated resection, it can be copied along with the adjacent cleavage-resistant sequence with great fidelity. The cleavage-resistant allele can either be naturally occurring or engineered (for example, to encode a synonymous codon). Because the gRNA sustaining copy-grafting will typically cut highly conserved wild-type alleles of the target sequence, such drives will replace both wild-type and undesired alleles with the preferred allele during the drive process. Despite its indirect action, copy-grafting can sustain drive at comparable frequencies to copy-cutting60.
Drive-neutralizing systems
Gene drives designed to alter the genetic architectures of populations have raised concerns regarding the potential unforeseen consequences of such engineering on the environment or disease transmission dynamics112,113,114,115,116. Two HDR-mediated gene-drive-neutralizing systems have been proposed68, modelled62,117,118 and tested62 that can either halt (erasing-CHACRs (e-CHACRs)) (Fig. 4d,e) or delete (ERACRs (elements reversing the autocatalytic chain reaction)) a gene drive (Fig. 4f,g). A key feature common to these two conditional drive systems is that they encode gRNAs but no source of Cas9. When the neutralizing elements encounter a gene drive, the Cas9 provided in trans by the gene drive combines with the gRNAs either to mutate and inactivate Cas9 (e-CHACRs), thereby preventing further drive, or to delete and replace the gene drive (ERACRs). In the absence of a Cas9 source, e-CHACRs and ERACRs are transmitted as simple Mendelian factors. However, when combined with a Cas9-powered gene drive, both neutralizing elements can copy themselves following logistic growth dynamics similar to gene-drive trajectories in wild-type populations.
e-CHACRs: active genetic elements that copy while mutating Cas9
e-CHACRs carry one gRNA to copy themselves at their site of genomic insertion and additional gRNAs to mutate and inactivate the Cas9 transgene68 (Fig. 4d). In Drosophila melanogaster62, all e-CHACRs analysed were highly efficient (>99%) in mutating and inactivating Cas9 with either of two different gRNAs that targeted sequences in the Cas9 transgene encoding catalytically essential amino acids. The Cas9-targeting gRNAs carried by e-CHACRs also sometimes induced local damage on the gene-drive chromosome, resulting in the production of homozygous lethal alleles and in a reduction in the prevalence of the gene drive. In the presence of Cas9, different e-CHACRs copied with a range of efficiencies (60–99% transmission). When an efficiently copying e-CHACR was challenged with a gene drive in cage experiments, it rapidly copied itself to homozygosity and eliminated Cas9 activity, providing proof of principle for this drive-neutralizing strategy62 (Fig. 4e).
ERACRs: active genetic elements that delete and replace gene drives
ERACRs are inserted into the genome at the same site as a gene-drive and carry two gRNAs that direct cleavage on either side of the drive62 (Fig. 4f) to delete the drive and copy ERACR sequences in its place. Although ERACRs often perform as designed, they also frequently deleted the drive without copying. The great majority of such deletion-only events damaged the target chromosome rendering it homozygous inviable. This type of local damage stems from each of the two gRNAs cutting at sites that are bordered on only one side by homologous sequences present on the target chromosome62, reminiscent of the damage associated with mutagenic transposition of the aberrantly nested Ds transposon element used in the classic studies of Barbara McClintock and analysed subsequently at the molecular level18,119.
ERACRs can also occasionally recombine with gene-drive sequences mediated by short (<20 bp) stretches of sequence complementarity62 via the synthesis-dependent strand annealing branch of the HDR repair pathway72,73,74,75. Notably, a few of these rare outcomes generate chimeric ERACR-drive elements that retain drive activity. Eliminating homology between ERACRs and gene-drives greatly reduced these infrequent events but they were still generated at low levels (<1%)62, perhaps by sporadic template switching during the repair process120.
Despite their imperfect performance, ERACRs largely performed as intended in population cage studies62 (Fig. 4f). An ERACR carrying recoded sequences that restored the activity of a gene whose requisite function was disrupted by insertion of the gene-drive rapidly replaced that drive element in population cages with remarkably little variation between cage replicates62 (Fig. 4g).
Another drive-neutralizing element, referred to as Cas9-triggered chain ablation (CATCHA)121, copies itself into the Cas9 transgene. The CATCHA element has design features similar to both the ERACR and e-CHACR in that it carries a gRNA targeting Cas9 for mutagenesis (like an e-CHACR) but is inserted into the same genomic site as the gene drive (like an ERACR). Alternatively, strains carrying functional drive-resistant alleles can potentially displace a gene drive that carries an appreciable fitness cost (for example, as suppression drives are designed to do71). Such cleavage-resistant alleles have the potential to outcompete the drive element as has been shown to occur in cage studies of early versions of suppression drives71,86. It should also be possible to combine these various drive-neutralizing approaches with elements carrying anti-Cas9 proteins. Indeed, recent results from the Crisanti group indicate that an anti-Cas9 transgene inherited in a Mendelian fashion can efficiently neutralize the activity of the suppression dsx-drive122. One intriguing possibility would be to equip a gene drive with an imperfect gRNA that inefficiently cleaves and inactivates Cas9 in a muted e-CHACR-like fashion62. Such a drive should spread with only minimally reduced kinetics but, after achieving full introduction, would mutagenize the Cas9 transgene at much higher rates than expected by spontaneous mutation. This type of self-limiting system should hasten elimination of Cas9 activity from a population if it incurred a significant fitness cost.
Developing drive systems in other organisms
Drive systems in mosquitoes and other insects could help combat additional devastating vector-borne diseases and control crop pests. In vertebrates, drives might immunize endangered species against specific pathogens123 (for example, protecting black-footed ferrets against the bubonic plague124) or re-establish native mammalian or bird species to aid in island conservation125,126,127. Conditional split active genetic systems might also accelerate aggregation of multiple desired traits, particularly in polyploid agricultural strains. For example, two or more gRNAs targeting sequences present on all allo-alleles could be edited in the presence of a Mendelian Cas9 source and then, following selfing, progeny could be identified carrying the multiple desired edits across all allo-alleles in the absence of the segregating Cas9 element. CRISPR-based drive-like systems, employing self-amplifying ‘pro-active genetic’ systems52 or RNA-guided transposons128,129,130,131,132, have also been developed in bacteria, which, if combined with a means for horizontal gene transfer133,134,135,136,137,138,139,140,141, could help deplete antibiotic resistance factors from bacteria in the environment142,143 and perhaps be adapted to fight bacterial infections138,139,140,141,144 (reviewed in ref.145).
Developing gene drives in other insect disease vectors and crop pests
The great advances in developing efficient gene drives in fruitflies and Anopheline mosquitoes46,47,48,54,84,85 offer hope that these technologies could be transferred to other mosquito species and diverse insect disease vectors, including other Anopheline species transmitting malaria146, arboviruses vectored by Ae. aegypti147, trypanosomes causing Chagas disease148 or sleeping sickness149 (transmitted by kissing bugs or Tsetse flies, respectively), Leishmaniasis spread by sand flies150, or West Nile virus151 and other encephalitic viruses152 infecting humans, birds and horses (vectored by Culex mosquitoes, for which site-directed transgenesis has just been reported153).
Although gene drives are likely to be adaptable to other insects, there may be considerable challenges in devising such systems, including colonizing new or native model mosquito species in the laboratory154,155, tuning methods for germline transgenesis to new insect species, or sustaining efficient gene conversion (for example, as has proven difficult in Ae. aegypti49). Simple practical matters can also impose barriers. For example, eggs of triatomine kissing bugs transmitting Chagas disease have extremely thick chorions (eggshells) that hinder DNA injection into germ cells156. The recent development of a method referred to as ‘ReMOT Control’ for delivering CRISPR tools into developing oocytes within adult ovaries of various insect species157,158,159,160 offers a promising alternative to egg injection. While this system has proven effective for ‘knockout’ gene editing, it remains to be determined whether it can deliver DNA repair templates to permit ‘knock-in’ modifications. These realistic considerations notwithstanding, solutions to overcome such technical barriers will most likely be developed in time with appropriate effort.
Gene-drive systems in mammals
Two prototype gene-drive systems developed in mammals50,161 could aid in efforts to rescue endangered species123 or to control invasive mammalian species and restore sensitive habitats to their native condition (for example, islands)125,126,127. The first is a split Cas9–gRNA drive similar in design to those developed in insects. The Cooper group inserted a gRNA-carrying drive cassette into the tyrosinase (Tyr) pigmentation locus in mice50 (Tyr–/– mutants have an albino phenotype) and combined this Tyr-drive element with a variety of Cas9 sources to determine which patterns of Cas9 expression (for example, ubiquitous or germline specific) might sustain drive (Fig. 5a). Although the results of this analysis reflect the differing reproductive biology of mammals versus insects, the overall lessons were much the same. Limiting Cas9 expression to the female germline resulted in substantial drive (reaching 78% transmission in one family); however, in contrast to insects, no copying was observed when Cas9 was expressed ubiquitously or restricted to male germline progenitor cells.
a | Although most gene-drive research has focused on insects, one path of current research is to expand gene-drive systems into other species, including mammalian species. The diagram shows a genetic map of a split-drive element inserted into the mouse tyrosinase (Tyr) locus50. When the element copies, the DsRed transgene it carries becomes associated with the chinchilla allelic variant on the target chromosome. b | Another key line of research is to translate insect drive systems from laboratory studies into field trials. Four phases of gene-drive development and implementation as delineated by the World Health Organization115,167. Phase 1 is laboratory development and testing. Phase 2 is confined outdoor tests (either in large cages or isolated environments such as islands); the goal of these experiments is to achieve defined entomological endpoints such as a certain percentage of population reduction (suppression) or drive introduction (modification). Phase 3 is limited testing in disease-endemic regions to demonstrate epidemiological efficacy. Phase 4 is full implementation. Community and regulatory engagement are conducted in parallel with all phases of this implementation plan. CMV, cytomegalovirus; gRNA, guide RNA; HDR, homology-directed repair; SNP, single nucleotide polymorphism. Part a is adapted from ref.50, Springer Nature Limited. Part b is adapted with permission from ref.69, Elsevier.
The common thread unifying successful mammalian and insect drives is restricting Cas9 expression to meiotic or pre-meiotic cells (pre-anaphase I) and to avoid earlier developmental expression in mitotically active cells. In mice, the female germline is allocated during embryogenesis and only a few mitotic cell cycles precede meiosis to generate post-replicative oocytes. By contrast, male germline cells undergo many mitotic divisions throughout adulthood, offering many more opportunities for NHEJ-induced alleles to be generated prior to meiotic phases where HDR-mediated copying of the drive element could occur. Future experiments optimizing female drive and restricting Cas9 expression to appropriate adult stages of male germline development may enable drive in both sexes and facilitate the design of suppression or modification strategies to achieve conservation goals.
The second mammalian drive concept is similar to that discussed above with regard to insect X-shredders but instead targets the Y chromosome for destruction161. The authors propose a two-component coupled-drive system in which a Cas9–gRNA-based drive located on an autosome is combined with an orthogonal CRISPR system, Cas12a (also known as Cpf1), which targets repeated sites on the Y chromosome. Modelling suggests that, if this Y-shredder were combined with an efficient autosomal drive, local elimination of an invasive target species as part of a bioremediation programme should be achievable so long as cleavage-resistant functional alleles at the drive locus did not arise.
Driving forwards
Progress in the gene-drive field has been remarkable over the past 5 years. In this brief period of intensive productivity, nearly all substantive technical barriers have been overcome for drive systems either modifying or suppressing mosquito populations. Based on these achievements, it is now possible to define relatively precise target–product profiles115,162 defining detailed characteristics of drive systems that would be suitable for advancing to the next phase of testing in physically or ecologically confined outdoor ‘phase 2’ field tests as defined by the World Health Organization phases of gene-drive development115. If drive systems perform similarly in these natural populations where mating success is highly competitive, they could be advanced to ‘phase 3’ trials in which the goal would be to demonstrate epidemiological efficacy in reducing the prevalence of malaria in a target area (Fig. 5b). Is this a realistic path forwards?
From the science and engineering perspective, the answer to the question seems likely to be yes (although one should not underestimate unforeseen challenges in nature). When considering the various social and ethical questions that have been raised regarding gene drives, the answer is less clear. An important consideration in this complex risk assessment is to balance the anticipated benefits of gene drives against their potential risks. Another issue concerns the longer-term consequences, both practical and ethical163,164,165,166, of going down the road of biologically engineering organisms in our environment. It will not be trivial to achieve a consensus answer to these deep questions.
I believe that a sensible format for evaluating the benefits versus risks of gene drives is to tally the real perceived benefits and risks of gene drives and then weigh and sum the various relevant factors164. It also makes sense to consider other well-known cases of naturally evolved gene-drive systems4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21 that have either driven themselves extinct or achieved some type of balanced equilibrium164.
With regard to the potential benefits of gene drives, malaria and other mosquito-borne diseases have been estimated by the Bill and Melinda Gates Foundation to kill more people than any other organism (including humans) and more than half of those deaths are caused by malaria. Although much progress has been made over the past decade in reducing the prevalence and intensity of malaria worldwide, the downward trend seems to have stalled owing to a variety of difficult challenges, including increased incidence of insecticide resistance in mosquitoes, increased drug resistance in parasites and maintenance of funding for these very costly efforts114,167. Global health challenges related to the current COVID-19 pandemic (and inevitable future disease outbreaks) will only aggravate these problems as vaccination programmes, access to drugs and basic health care become strained.
Summary and conclusions
Since getting on the road, gene-drives have sped a long way. New high-performance vehicles offer great promise for delivering anti-malarial effectors or driving down the numbers of mosquitoes. Flexible add-on trailers (for example, CHACRs) could also update or expand the range of the original drives and, should a drive go errant, strategies for forcing them to pull-over (e-CHACRs) or exit (ERACRs) are now available. Such technologies should be portable to other insects and, with further development, perhaps transferred to other organisms, including vertebrates, plants and even bacteria. So, what is next?
Beyond the several technical and ethical challenges described in this Review and elsewhere, I would like to highlight two main challenges facing the gene-drive field. The first is to obtain both regulatory and community approval to test these systems in natural confined environments such as isolated islands (phase 2 trials) or other controlled contexts. These trials are essential for obtaining data to evaluate the future potential of candidate drive systems. There is no other way to know how drives will perform in nature under conditions where they must compete with native mosquitoes, which may be much more extreme than in the laboratory. Approval for such phase 2 trials may take time and will depend greatly on efforts such as those already well under way to engage local communities in an open and transparent fashion regarding scientific and ethical issues163,166. The second challenge, which in many ways rests on the results of the first, is to delineate when and where such drives could be released in phase 3 efforts to reduce disease prevalence. Each drive system will need to develop a detailed target–product profile, which should be evaluated on a case-by-case basis. It will also be important to consider how new drive systems might interact with those already in the environment to avoid potential clashes analogous to those arising from space junk.
In summary, so far, it has been a remarkable drive, through an incredible landscape. The big question is what lies on the road ahead and will the promise of gene-drives deliver the potential they seem to hold?
References
Serebrovsky, A. S. On the possibility of a new method for the control of insect pests. Zool. Zh. 19, 618–630 (1940).
Curtis, C. F. Possible use of translocations to fix desirable genes in insect pest populations. Nature 218, 368–369 (1968). This paper is one of the first to describe how reciprocal chromosomal translocations could be used to drive a favoured linked trait in a threshold-dependent fashion.
Dawkins, R. The Selfish Gene Vol. 345 (Oxford University Press, 1976).
Bastide, H. et al. Rapid rise and fall of selfish sex-ratio X chromosomes in Drosophila simulans: spatiotemporal analysis of phenotypic and molecular data. Mol. Biol. Evol. 28, 2461–2470 (2011).
Corbett-Detig, R., Medina, P., Frerot, H., Blassiau, C. & Castric, V. Bulk pollen sequencing reveals rapid evolution of segregation distortion in the male germline of Arabidopsis hybrids. Evol. Lett. 3, 93–103 (2019).
Kingan, S. B., Garrigan, D. & Hartl, D. L. Recurrent selection on the Winters sex-ratio genes in Drosophila simulans. Genetics 184, 253–265 (2010).
McLaughlin, R. N. Jr. & Malik, H. S. Genetic conflicts: the usual suspects and beyond. J. Exp. Biol. 220, 6–17 (2017).
Presgraves, D. C., Gerard, P. R., Cherukuri, A. & Lyttle, T. W. Large-scale selective sweep among segregation distorter chromosomes in African populations of Drosophila melanogaster. PLoS Genet. 5, e1000463 (2009).
Seymour, D. K., Chae, E., Arioz, B. I., Koenig, D. & Weigel, D. Transmission ratio distortion is frequent in Arabidopsis thaliana controlled crosses. Heredity 122, 294–304 (2019).
Courret, C., Chang, C. H., Wei, K. H., Montchamp-Moreau, C. & Larracuente, A. M. Meiotic drive mechanisms: lessons from Drosophila. Proc. Biol. Sci. 286, 20191430 (2019).
Kusano, A., Staber, C., Chan, H. Y. & Ganetzky, B. Closing the (Ran)GAP on segregation distortion in Drosophila. Bioessays 25, 108–115 (2003).
Merel, V., Boulesteix, M., Fablet, M. & Vieira, C. Transposable elements in Drosophila. Mob. DNA 11, 23 (2020).
Boulesteix, M. & Biemont, C. Transposable elements in mosquitoes. Cytogenet. Genome Res. 110, 500–509 (2005).
Lee, Y. C. & Langley, C. H. Transposable elements in natural populations of Drosophila melanogaster. Philos. Trans. R. Soc. Lond. B Biol. Sci. 365, 1219–1228 (2010).
Kelleher, E. S. Reexamining the P-element invasion of Drosophila melanogaster through the lens of piRNA silencing. Genetics 203, 1513–1531 (2016).
Majumdar, S. & Rio, D. C. P transposable elements in drosophila and other eukaryotic organisms. Microbiol. Spectr. 3, MDNA3–0004-2014 (2015).
Burns, K. H. & Boeke, J. D. Human transposon tectonics. Cell 149, 740–752 (2012).
Doring, H. P., Tillmann, E. & Starlinger, P. DNA sequence of the maize transposable element Dissociation. Nature 307, 127–130 (1984).
Wallau, G. L., Capy, P., Loreto, E. & Hua-Van, A. Genomic landscape and evolutionary dynamics of mariner transposable elements within the Drosophila genus. BMC Genomics 15, 727 (2014).
Hawkins, J. S., Hu, G., Rapp, R. A., Grafenberg, J. L. & Wendel, J. F. Phylogenetic determination of the pace of transposable element proliferation in plants: copia and LINE-like elements in Gossypium. Genome 51, 11–18 (2008).
Biemont, C., Vieira, C., Borie, N. & Lepetit, D. Transposable elements and genome evolution: the case of Drosophila simulans. Genetica 107, 113–120 (1999).
Buchman, A. B., Ivy, T., Marshall, J. M., Akbari, O. S. & Hay, B. A. Engineered reciprocal chromosome translocations drive high threshold, reversible population replacement in drosophila. ACS Synth. Biol. 7, 1359–1370 (2018).
Akbari, O. S. et al. Novel synthetic Medea selfish genetic elements drive population replacement in Drosophila; a theoretical exploration of Medea-dependent population suppression. ACS Synth. Biol. 3, 915–928 (2014).
Buchman, A., Marshall, J. M., Ostrovski, D., Yang, T. & Akbari, O. S. Synthetically engineered Medea gene drive system in the worldwide crop pest Drosophila suzukii. Proc. Natl Acad. Sci. USA 115, 4725–4730 (2018).
Champer, J., Zhao, J., Champer, S. E., Liu, J. & Messer, P. W. Population dynamics of underdominance gene drive systems in continuous space. ACS Synth. Biol. 9, 779–792 (2020).
Chen, C. C. et al. EXO1 suppresses double-strand break induced homologous recombination between diverged sequences in mammalian cells. DNA Repair. 57, 98–106 (2017).
Leftwich, P. T. et al. Recent advances in threshold-dependent gene drives for mosquitoes. Biochem. Soc. Trans. 46, 1203–1212 (2018).
Raban, R. R., Marshall, J. M. & Akbari, O. S. Progress towards engineering gene drives for population control. J. Exp. Biol. 223 (Suppl. 1), jeb208181 (2020).
Ward, C. M. et al. Medea selfish genetic elements as tools for altering traits of wild populations: a theoretical analysis. Evolution 65, 1149–1162 (2011).
Oberhofer, G., Ivy, T. & Hay, B. A. Gene drive and resilience through renewal with next generation Cleave and Rescue selfish genetic elements. Proc. Natl Acad. Sci. USA 117, 9013–9021 (2020).
Oberhofer, G., Ivy, T. & Hay, B. A. Cleave and Rescue, a novel selfish genetic element and general strategy for gene drive. Proc. Natl Acad. Sci. USA 116, 6250–6259 (2019).
Champer, J. et al. A toxin-antidote CRISPR gene drive system for regional population modification. Nat. Commun. 11, 1082 (2020).
Yen, P. S. & Failloux, A. B. A review: Wolbachia-based population replacement for mosquito control shares common points with genetically modified control approaches. Pathogens 9, 404 (2020).
O’Neill, S. L. The use of wolbachia by the world mosquito program to interrupt transmission of aedes aegypti transmitted viruses. Adv. Exp. Med. Biol. 1062, 355–360 (2018).
Niang, E. H. A., Bassene, H., Fenollar, F. & Mediannikov, O. Biological control of mosquito-borne diseases: the potential of wolbachia-based interventions in an IVM framework. J. Trop. Med. 2018, 1470459 (2018).
Chevalier, B. S. & Stoddard, B. L. Homing endonucleases: structural and functional insight into the catalysts of intron/intein mobility. Nucleic Acids Res. 29, 3757–3774 (2001).
Macreadie, I. G., Scott, R. M., Zinn, A. R. & Butow, R. A. Transposition of an intron in yeast mitochondria requires a protein encoded by that intron. Cell 41, 395–402 (1985).
Rong, Y. S. & Golic, K. G. The homologous chromosome is an effective template for the repair of mitotic DNA double-strand breaks in Drosophila. Genetics 165, 1831–1842 (2003).
Chan, Y. S., Huen, D. S., Glauert, R., Whiteway, E. & Russell, S. Optimising homing endonuclease gene drive performance in a semi-refractory species: the Drosophila melanogaster experience. PLoS ONE 8, e54130 (2013).
Windbichler, N. et al. A synthetic homing endonuclease-based gene drive system in the human malaria mosquito. Nature 473, 212–215 (2011). This study is the first demonstration of nuclease-mediated gene drive in mosquitoes based on the homing endonuclease gene I-SceI.
Carroll, D. Genome engineering with targetable nucleases. Annu. Rev. Biochem. 83, 409–439 (2014).
Barrangou, R. et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712 (2007).
Jinek, M. et al. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821 (2012). This foundational study developed the most widely used dual synthetic CRISPR system consisting of Cas9 endonuclease and gRNA components.
Doudna, J. A., Sternberg, S. H. A Crack in Creation: Gene Editing and the Unthinkable Power to Control Evolution 281 (Houghton Mifflin Harcourt, 2017).
Gantz, V. M. & Bier, E. The mutagenic chain reaction: a method for converting heterozygous to homozygous mutations. Science 348, 442–444 (2015). This study reported the first CRISPR-based gene drive in a metazoan organism (D. melanogaster) with a specialized germline.
Gantz, V. M. et al. Highly efficient Cas9-mediated gene drive for population modification of the malaria vector mosquito Anopheles stephensi. Proc. Natl Acad. Sci. USA 112, E6736–E6743 (2015). This study describes the first efficient CRISPR-based gene drive system in mosquitoes, which carried a dual anti-malarial effector cassette.
Hammond, A. et al. A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nat. Biotechnol. 34, 78–83 (2016). This study describes the first efficient CRISPR-based suppression gene drive system in mosquitoes.
Kyrou, K. et al. A CRISPR-Cas9 gene drive targeting doublesex causes complete population suppression in caged Anopheles gambiae mosquitoes. Nat. Biotechnol. 36, 1062–1066 (2018). This study describes a highly efficient suppression gene drive system in mosquitoes targeting an invariant genome target site in the doublesex locus.
Li, M. et al. Development of a confinable gene drive system in the human disease vector Aedes aegypti. eLife 9, e51701 (2020).
Grunwald, H. A. et al. Super-Mendelian inheritance mediated by CRISPR-Cas9 in the female mouse germline. Nature 566, 105–109 (2019). This study provided the first proof-of-principle gene drive system in mammals, which selectively sustained drive via the female germline.
DiCarlo, J. E., Chavez, A., Dietz, S. L., Esvelt, K. M. & Church, G. M. Safeguarding CRISPR-Cas9 gene drives in yeast. Nat. Biotechnol. 33, 1250–1255 (2015). This study demonstrated CRISPR-based gene conversion in diploid yeast, which could then be transmitted meiotically.
Valderrama, J. A., Kulkarni, S. S., Nizet, V. & Bier, E. A bacterial gene-drive system efficiently edits and inactivates a high copy number antibiotic resistance locus. Nat. Commun. 10, 5726 (2019). This study generalizes the concept of gene drive to bacteria, where it is applied to efficiently reduce the frequency of antibiotic reistance.
Esvelt, K. M., Smidler, A. L., Catteruccia, F. & Church, G. M. Concerning RNA-guided gene drives for the alteration of wild populations. eLife 3, e03401 (2014).
Adolfi, A. et al. Efficient population modification gene-drive rescue system in the malaria mosquito Anopheles stephensi. Nat. Commun. 11, 5553 (2020). This study reports on the first recoded gene drive in mosquitoes that drove efficiently through both males and females based on the process of lethal/sterile mosaicism.
Champer, J. et al. A CRISPR homing gene drive targeting a haplolethal gene removes resistance alleles and successfully spreads through a cage population. Proc. Natl Acad. Sci. USA 117, 24377–24383 (2020).
Kandul, N. P., Liu, J., Bennett, J. B., Marshall, J. M. & Akbari, O. S. A confinable home-and-rescue gene drive for population modification. eLife 10, e65939 (2021).
Terradas, G. et al. Inherently confinable split-drive systems in Drosophila. Nat. Commun. 12, 1480 (2021). This study further develops the strategy of inserting a recoded gene drive in genes essential for viability or reproduction in the context of split drive systems.
Xu, X. S., Gantz, V. M., Siomava, N. & Bier, E. CRISPR/Cas9 and active genetics-based trans-species replacement of the endogenous Drosophila kni-L2 CRM reveals unexpected complexity. eLife 6, e30281 (2017).
Lopez Del Amo, V. et al. A transcomplementing gene drive provides a flexible platform for laboratory investigation and potential field deployment. Nat. Commun. 11, 352 (2020). This study reports on the reconstitution of a full gene drive from split constituent parts.
Guichard, A. et al. Efficient allelic-drive in Drosophila. Nat. Commun. 10, 1640 (2019). The study develops two allelic drive systems, copy-cutting and copy-grafting, to propagate favoured alleles of an essential gene.
Kandul, N. P. et al. Assessment of a split homing based gene drive for efficient knockout of multiple genes. G3 10, 827–837 (2020).
Xu, X.-R. S. et al. Active-genetic neutralizing elements for halting or deleting gene-drives. Mol. Cell 80, 246–262 (2020). This study reports on two drive-neutralizing systems that either inactivate (e-CHACR) or delete and replace (ERACR) a gene drive.
Burt, A. Site-specific selfish genes as tools for the control and genetic engineering of natural populations. Proc. Biol. Sci. 270, 921–928 (2003). This seminal modelling study provides the theoretical underpinnings for the modern gene-drive field.
North, A. R., Burt, A. & Godfray, H. C. J. Modelling the potential of genetic control of malaria mosquitoes at national scale. BMC Biol. 17, 26 (2019). This study provides a comprehensive analysis of the perfomance of suppressive gene drives following iterative releases across various topographies.
North, A. R., Burt, A. & Godfray, H. C. J. Modelling the suppression of a malaria vector using a CRISPR-Cas9 gene drive to reduce female fertility. BMC Biol. 18, 98 (2020).
Collins, C. M., Bonds, J. A. S., Quinlan, M. M. & Mumford, J. D. Effects of the removal or reduction in density of the malaria mosquito, Anopheles gambiae s.l., on interacting predators and competitors in local ecosystems. Med. Vet. Entomol. 33, 1–15 (2019).
James, A. A. Gene drive systems in mosquitoes: rules of the road. Trends Parasitol. 21, 64–67 (2005).
Gantz, V. M. & Bier, E. The dawn of active genetics. Bioessays 38, 50–63 (2016).
Macias, V. M. & James, A. A. in Genetic Control of Malaria and Dengue (ed. Adelman, Z. N.) 423–444 (Elsevier Academic Press, 2015).
Eckhoff, P. A., Wenger, E. A., Godfray, H. C. & Burt, A. Impact of mosquito gene drive on malaria elimination in a computational model with explicit spatial and temporal dynamics. Proc. Natl Acad. Sci. USA 114, E255–E264 (2017). This study provides a detailed analysis of drive parameters relevant to both suppression-based and modification-based drives and is the first to model a drive in the context of a two-dimensional environment.
Hammond, A. M. et al. The creation and selection of mutations resistant to a gene drive over multiple generations in the malaria mosquito. PLoS Genet. 13, e1007039 (2017).
Joyce, E. F., Paul, A., Chen, K. E., Tanneti, N. & McKim, K. S. Multiple barriers to nonhomologous DNA end joining during meiosis in Drosophila. Genetics 191, 739–746 (2012).
Bozas, A., Beumer, K. J., Trautman, J. K. & Carroll, D. Genetic analysis of zinc-finger nuclease-induced gene targeting in Drosophila. Genetics 182, 641–651 (2009).
Do, A. T., Brooks, J. T., Le Neveu, M. K. & LaRocque, J. R. Double-strand break repair assays determine pathway choice and structure of gene conversion events in Drosophila melanogaster. G3 4, 425–432 (2014).
Wei, D. S. & Rong, Y. S. A genetic screen for DNA double-strand break repair mutations in Drosophila. Genetics 177, 63–77 (2007).
Lin, C. C. & Potter, C. J. Non-Mendelian dominant maternal effects caused by CRISPR/Cas9 transgenic components in Drosophila melanogaster. G3 6, 3685–3691 (2016).
Champer, J. et al. Novel CRISPR/Cas9 gene drive constructs reveal insights into mechanisms of resistance allele formation and drive efficiency in genetically diverse populations. PLoS Genet. 13, e1006796 (2017).
Anopheles gambiae 1000 Genomes Consortiumet al. Genetic diversity of the African malaria vector Anopheles gambiae. Nature 552, 96–100 (2017).
Deredec, A., Burt, A. & Godfray, H. C. The population genetics of using homing endonuclease genes in vector and pest management. Genetics 179, 2013–2026 (2008).
Fasulo, B. et al. A fly model establishes distinct mechanisms for synthetic CRISPR/Cas9 sex distorters. PLoS Genet. 16, e1008647 (2020).
Galizi, R. et al. A synthetic sex ratio distortion system for the control of the human malaria mosquito. Nat. Commun. 5, 3977 (2014).
Galizi, R. et al. A CRISPR-Cas9 sex-ratio distortion system for genetic control. Sci. Rep. 6, 31139 (2016).
Turner, J. M. Meiotic sex chromosome inactivation. Development 134, 1823–1831 (2007).
Simoni, A. et al. A male-biased sex-distorter gene drive for the human malaria vector Anopheles gambiae. Nat. Biotechnol. 38, 1054–1060 (2020).
Carballar-Lejarazu, R. & et al. Next-generation gene drive for population modification of the malaria vector mosquito, Anopheles gambiae.Proc. Natl Acad. Sci. USA 117, 22805–22814 (2020). This study describes a modification gene drive that propagates with high efficiency through both males and females.
Pham, T. B. et al. Experimental population modification of the malaria vector mosquito, Anopheles stephensi. PLoS Genet. 15, e1008440 (2019).
Dong, Y., Simoes, M. L. & Dimopoulos, G. Versatile transgenic multistage effector-gene combinations for Plasmodium falciparum suppression in Anopheles. Sci. Adv. 6, eaay5898 (2020).
Dong, Y. et al. Engineered anopheles immunity to Plasmodium infection. PLoS Pathog. 7, e1002458 (2011).
Isaacs, A. T. et al. Transgenic Anopheles stephensi coexpressing single-chain antibodies resist Plasmodium falciparum development. Proc. Natl Acad. Sci. USA 109, E1922–E1930 (2012). This study demonstrates 100% protection against parasite transmission in transgenic mosquitoes carrying a dual anti-parasite effector cassette.
Haber, J. E. TOPping off meiosis. Mol. Cell 57, 577–581 (2015).
Hammond, A. et al. Regulating the expression of gene drives is key to increasing their invasive potential and the mitigation of resistance. PLoS Genet. 17, e1009321 (2021).
Lee, Y. et al. Genome-wide divergence among invasive populations of Aedes aegypti in California. BMC Genomics 20, 204 (2019).
Callaway, E. Gene drives thwarted by emergence of resistant organisms. Nature 542, 15 (2017).
Unckless, R. L., Clark, A. G. & Messer, P. W. Evolution of resistance against CRISPR/Cas9 gene drive. Genetics 205, 827–841 (2017).
Drury, D. W., Dapper, A. L., Siniard, D. J., Zentner, G. E. & Wade, M. J. CRISPR/Cas9 gene drives in genetically variable and nonrandomly mating wild populations. Sci. Adv. 3, e1601910 (2017).
Schmidt, H. et al. Abundance of conserved CRISPR-Cas9 target sites within the highly polymorphic genomes of Anopheles and Aedes mosquitoes. Nat. Commun. 11, 1425 (2020). This study provides computational evidence that conserved CRISPR cleavage sites are abundant in the genome.
Akbari, O. S. et al. Safeguarding gene drive experiments in the laboratory. Science 349, 927–929 (2015).
Li, J. et al. Genome-block expression-assisted association studies discover malaria resistance genes in Anopheles gambiae. Proc. Natl Acad. Sci. USA 110, 20675–20680 (2013).
Niu, G. et al. The fibrinogen-like domain of FREP1 protein is a broad-spectrum malaria transmission-blocking vaccine antigen. J. Biol. Chem. 292, 11960–11969 (2017).
Zhang, G. et al. Anopheles midgut FREP1 mediates plasmodium invasion. J. Biol. Chem. 290, 16490–16501 (2015).
Dong, Y., Simoes, M. L., Marois, E. & Dimopoulos, G. CRISPR/Cas9 -mediated gene knockout of Anopheles gambiae FREP1 suppresses malaria parasite infection. PLoS Pathog. 14, e1006898 (2018).
Simoes, M. L., Caragata, E. P. & Dimopoulos, G. Diverse host and restriction factors regulate mosquito-pathogen interactions. Trends Parasitol. 34, 603–616 (2018).
Nash, A. et al. Integral gene drives for population replacement. Biol. Open 8, bio037762 (2019). This study describes a bipartite drive system that can enable testing of anti-parasite effector cassettes under standard mosquito confinement protocols.
Enayati, A., Hanafi-Bojd, A. A., Sedaghat, M. M., Zaim, M. & Hemingway, J. Evolution of insecticide resistance and its mechanisms in Anopheles stephensi in the WHO Eastern Mediterranean Region. Malar. J. 19, 258 (2020).
Ffrench-Constant, R. H., Williamson, M. S., Davies, T. G. & Bass, C. Ion channels as insecticide targets. J. Neurogenet. 30, 163–177 (2016).
Silva, J. J. & Scott, J. G. Conservation of the voltage-sensitive sodium channel protein within the Insecta. Insect Mol. Biol. 29, 9–18 (2020).
Casida, J. E. & Durkin, K. A. Novel GABA receptor pesticide targets. Pestic. Biochem. Physiol. 121, 22–30 (2015).
Ihara, M., Buckingham, S. D., Matsuda, K. & Sattelle, D. B. Modes of action, resistance and toxicity of insecticides targeting nicotinic acetylcholine receptors. Curr. Med. Chem. 24, 2925–2934 (2017).
Thapa, S., Lv, M. & Xu, H. Acetylcholinesterase: a primary target for drugs and insecticides. Mini Rev. Med. Chem. 17, 1665–1676 (2017).
Kleinstiver, B. P. et al. Engineered CRISPR-Cas12a variants with increased activities and improved targeting ranges for gene, epigenetic and base editing. Nat. Biotechnol. 37, 276–282 (2019).
Walton, R. T., Christie, K. A., Whittaker, M. N. & Kleinstiver, B. P. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science 368, 290–296 (2020).
Committee on Gene Drive Research in Non-Human Organisms: Recommendations for Responsible Conduct; Board on Life Sciences; Division on Earth and Life Studies; National Academies of Sciences, Engineering, and Medicine. Gene Drives on the Horizon: Advancing Science, Navigating Uncertainty, and Aligning Research with Public Values (The National Academies Press, 2016). This comprehensive advisory and historical review document summarizes consensus views for how to safely rear and study gene-drive systems in the laboratory.
Adelman, Z. et al. Rules of the road for insect gene drive research and testing. Nat. Biotechnol. 35, 716–718 (2017).
James, S. et al. Pathway to deployment of gene drive mosquitoes as a potential biocontrol tool for elimination of malaria in Sub-Saharan Africa: recommendations of a scientific working group(dagger). Am. J. Trop. Med. Hyg. 98, 1–49 (2018).
James, S. L., Marshall, J. M., Christophides, G. K., Okumu, F. O. & Nolan, T. Toward the definition of efficacy and safety criteria for advancing gene drive-modified mosquitoes to field testing. Vector Borne Zoonotic Dis. 20, 237–251 (2020).
Warmbrod, K. L. et al. Gene Drives: Pursuing Opportunities, Minimizing Risk - A Johns Hopkins University Report on Responsible Governance (Johns Hopkins Bloomberg School of Public Health, Center for Health Security, Johns Hopkins University, 2020).
Vella, M. R., Gunning, C. E., Lloyd, A. L. & Gould, F. Evaluating strategies for reversing CRISPR-Cas9 gene drives. Sci. Rep. 7, 11038 (2017).
Rode, N. O., Courtier-Orgogozo, V. & Debarre, F. Can a population targeted by a CRISPR-based homing gene drive be rescued? G3 10, 3403–3415 (2020).
Fedoroff, N., Wessler, S. & Shure, M. Isolation of the transposable maize controlling elements Ac and Ds. Cell 35, 235–242 (1983).
Paix, A. et al. Precision genome editing using synthesis-dependent repair of Cas9-induced DNA breaks. Proc. Natl Acad. Sci. USA 114, E10745–E10754 (2017).
Wu, B., Luo, L. & Gao, X. J. Cas9-triggered chain ablation of cas9 as a gene drive brake. Nat. Biotechnol. 34, 137–138 (2016).
Taxiarchi, C. et al. A genetically encoded anti-CRISPR protein constrains gene drive spread and prevents population suppression. Nat. Commun. 12, 3977 (2021).
Conklin, B. R. On the road to a gene drive in mammals. Nature 566, 43–45 (2019).
Salkeld, D. J. Vaccines for conservation: plague, prairie dogs & black-footed ferrets as a case study. Ecohealth 14, 432–437 (2017).
Teem, J. L. et al. Genetic biocontrol for invasive species. Front. Bioeng. Biotechnol. 8, 452 (2020).
Godwin, J. et al. Rodent gene drives for conservation: opportunities and data needs. Proc. Biol. Sci. 286, 20191606 (2019).
McFarlane, G. R., Whitelaw, C. B. A. & Lillico, S. G. CRISPR-based gene drives for pest control. Trends Biotechnol. 36, 130–133 (2018).
Klompe, S. E., Vo, P. L. H., Halpin-Healy, T. S. & Sternberg, S. H. Transposon-encoded CRISPR-Cas systems direct RNA-guided DNA integration. Nature 571, 219–225 (2019).
Koonin, E. V., Makarova, K. S., Wolf, Y. I. & Krupovic, M. Evolutionary entanglement of mobile genetic elements and host defence systems: guns for hire. Nat. Rev. Genet. 21, 119–131 (2020).
Strecker, J. et al. RNA-guided DNA insertion with CRISPR-associated transposases. Science 365, 48–53 (2019).
Peters, J. E., Makarova, K. S., Shmakov, S. & Koonin, E. V. Recruitment of CRISPR-Cas systems by Tn7-like transposons. Proc. Natl Acad. Sci. USA 114, E7358–E7366 (2017).
Wiegand, T. & Wiedenheft, B. CRISPR Surveillance Turns Transposon Taxi. CRISPR J. 3, 10–12 (2020).
Hamilton, T. A. et al. Efficient inter-species conjugative transfer of a CRISPR nuclease for targeted bacterial killing. Nat. Commun. 10, 4544 (2019).
Price, V. J. et al. Enterococcus faecalis CRISPR-cas is a robust barrier to conjugative antibiotic resistance dissemination in the murine intestine. mSphere 4, e00464-19 (2019).
Rodrigues, M., McBride, S. W., Hullahalli, K., Palmer, K. L. & Duerkop, B. A. Conjugative delivery of CRISPR-Cas9 for the selective depletion of antibiotic-resistant enterococci. Antimicrob. Agents Chemother. 63, e01454-19 (2019).
Carraro, N. et al. Plasmid-like replication of a minimal streptococcal integrative and conjugative element. Microbiology 162, 622–632 (2016).
Brophy, J. A. N. et al. Engineered integrative and conjugative elements for efficient and inducible DNA transfer to undomesticated bacteria. Nat. Microbiol. 3, 1043–1053 (2018).
Bikard, D. et al. Exploiting CRISPR-Cas nucleases to produce sequence-specific antimicrobials. Nat. Biotechnol. 32, 1146–1150 (2014).
Citorik, R. J., Mimee, M. & Lu, T. K. Sequence-specific antimicrobials using efficiently delivered RNA-guided nucleases. Nat. Biotechnol. 32, 1141–1145 (2014).
Yosef, I., Manor, M., Kiro, R. & Qimron, U. Temperate and lytic bacteriophages programmed to sensitize and kill antibiotic-resistant bacteria. Proc. Natl Acad. Sci. USA 112, 7267–7272 (2015).
Park, J. Y. et al. Genetic engineering of a temperate phage-based delivery system for CRISPR/Cas9 antimicrobials against Staphylococcus aureus. Sci. Rep. 7, 44929 (2017).
Pazda, M., Kumirska, J., Stepnowski, P. & Mulkiewicz, E. Antibiotic resistance genes identified in wastewater treatment plant systems - a review. Sci. Total. Env. 697, 134023 (2019).
Kraemer, S. A., Ramachandran, A. & Perron, G. G. Antibiotic pollution in the environment: from microbial ecology to public policy. Microorganisms 7, 180 (2019).
Ram, G., Ross, H. F., Novick, R. P., Rodriguez-Pagan, I. & Jiang, D. Conversion of staphylococcal pathogenicity islands to CRISPR-carrying antibacterial agents that cure infections in mice. Nat. Biotechnol. 36, 971–976 (2018).
Bier, E. & Nizet, V. Driving to safety: CRISPR-based genetic approaches to reducing antibiotic resistance. Trends Genet. https://doi.org/10.1016/j.tig.2021.02.007 (2021).
Rossati, A. et al. Climate, environment and transmission of malaria. Infez. Med. 24, 93–104 (2016).
Fontenille, D. & Powell, J. R. From anonymous to public enemy: how does a mosquito become a feared arbovirus vector? Pathogens 9, 265 (2020).
Lidani, K. C. F. et al. Chagas disease: from discovery to a worldwide health problem. Front. Public Health 7, 166 (2019).
Buscher, P., Cecchi, G., Jamonneau, V. & Priotto, G. Human African trypanosomiasis. Lancet 390, 2397–2409 (2017).
Desjeux, P. Leishmaniasis: current situation and new perspectives. Comp. Immunol. Microbiol. Infect. Dis. 27, 305–318 (2004).
Saxena, V., Bolling, B. G. & Wang, T. West nile virus. Clin. Lab. Med. 37, 243–252 (2017).
Simon, L. V., Kong, E. L. & Graham, C. in St. Louis Encephalitis (StatPearls, 2020).
Feng, X. et al. Optimized CRISPR tools and site-directed transgenesis towards gene drive development in Culex quinquefasciatus mosquitoes. Nat. Commun. 12, 2960 (2021).
Nepomichene, T. N., Andrianaivolambo, L., Boyer, S. & Bourgouin, C. Efficient method for establishing F1 progeny from wild populations of Anopheles mosquitoes. Malar. J. 16, 21 (2017).
Marchand, R. P. A new cage for observing mating behavior of wild Anopheles gambiae in the laboratory. J. Am. Mosq. Control. Assoc. 1, 234–236 (1985).
Nunes-da-Fonseca, R., Berni, M., Tobias-Santos, V., Pane, A. & Araujo, H. M. Rhodnius prolixus: from classical physiology to modern developmental biology. Genesis https://doi.org/10.1002/dvg.22995 (2017).
Chaverra-Rodriguez, D. et al. Targeted delivery of CRISPR-Cas9 ribonucleoprotein into arthropod ovaries for heritable germline gene editing. Nat. Commun. 9, 3008 (2018).
Macias, V. M. et al. Cas9-mediated gene-editing in the malaria mosquito anopheles stephensi by ReMOT Control. G3 10, 1353–1360 (2020).
Chaverra-Rodriguez, D. et al. Germline mutagenesis of Nasonia vitripennis through ovarian delivery of CRISPR-Cas9 ribonucleoprotein. Insect Mol. Biol. 29, 569–577 (2020).
Heu, C. C., McCullough, F. M., Luan, J. & Rasgon, J. L. CRISPR-Cas9-based genome editing in the silverleaf whitefly (Bemisia tabaci). CRISPR J. 3, 89–96 (2020).
Prowse, T. A., Adikusuma, F., Cassey, P., Thomas, P. & Ross, J. V. A Y-chromosome shredding gene drive for controlling pest vertebrate populations. eLife 8, e41873 (2019).
Carballar-Lejarazu, R. & James, A. A. Population modification of Anopheline species to control malaria transmission. Pathog. Glob. Health 111, 424–435 (2017).
Annas, G. J. et al. A code of ethics for gene drive research. CRISPR J. 4, 19–24 (2021).
Bier, E. & Sober, E. Gene editing and the war against malaria. Am. Sci. 108, 162–169 (2020).
Long, K. C. et al. Core commitments for field trials of gene drive organisms. Science 370, 1417–1419 (2020).
Kormos, A. et al. Application of the relationship-based model to engagement for field trials of genetically engineered malaria vectors.Am. J. Trop. Med. Hyg. 104, 805–811 (2020).
World Health Organization. Guidance framework for testing of genetically modified mosquitoes. WHO http://apps.who.int/iris/bitstream/10665/127889/1/9789241507486_eng.pdf (2014).
Smith, D. L., McKenzie, F. E., Snow, R. W. & Hay, S. I. Revisiting the basic reproductive number for malaria and its implications for malaria control. PLoS Biol. 5, e42 (2007).
Brauer, F., Castillo-Chavez, C., Mubayi, A. & Towers, S. Some models for epidemics of vector-transmitted diseases. Infect. Dis. Model. 1, 79–87 (2016).
Deredec, A., Godfray, H. C. & Burt, A. Requirements for effective malaria control with homing endonuclease genes. Proc. Natl Acad. Sci. USA 108, E874–E880 (2011).
Escalante, A. A. & Pacheco, M. A. Malaria molecular epidemiology: an evolutionary genetics perspective. Microbiol. Spectr. https://doi.org/10.1128/microbiolspec.AME-0010-2019 (2019).
Selvaraj, P. et al. Vector genetics, insecticide resistance and gene drives: An agent-based modeling approach to evaluate malaria transmission and elimination. PLoS Comput. Biol. 16, e1008121 (2020).
Acknowledgements
These studies were supported by NIH grant R01 GM117321, a Paul G. Allen Frontiers Group Distinguished Investigator Award to E.B., the Bill and Melinda Gates Foundation and a gift from the Tata Trusts in India to TIGS-UCSD. The author thanks his UCI-Malaria Initiative colleagues: A. A. James, G. Dimopoulos, G. Lanzaro, A. Kormos, and J. Marshall for thoughtful discussions and comments on the manuscript as well as A. A. James and A. Crisanti for granting permission to cite some of their unpublished results.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
E.B. has equity interest in two companies: Synbal Inc. and Agragene, Inc. These companies may potentially benefit from the research results. E.B. also serves on Synbal Inc.’s Board of Directors and Scientific Advisory Board and on Agragene Inc.’s Scientific Advisory Board. The terms of these arrangements have been reviewed and approved by the University of California, San Diego, in accordance with its conflict of interest policies.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
FNIH GeneConvene Global Collaborative: https://fnih.org/our-programs/geneconvene
PlasmoDB - Plasmodium Informatics Resources: https://plasmodb.org/plasmo/app
Target Malaria: https://targetmalaria.org/
TIGS: Tata Institutes for Genetics and Society, UCSD: https://tigs.ucsd.edu/
UCIMI: University of California Irvine Malaria Initiative: https://ucimi.org/
WHO Guidelines for malaria: https://app.magicapp.org/#/guideline/5438
WHO Malaria: https://www.who.int/news-room/fact-sheets/detail/malaria
WHO World Malaria Report 2019: https://www.who.int/publications/i/item/9789241565721
Supplementary information
Glossary
- Gene drives
-
An allele of a diploid gene experiences gene drive if it is inherited more than 50% of the time (that is, more than by random chance).
- High-threshold drives
-
Drive systems wherein release of many individuals is required for super-Mendelian spread of the drive.
- Low-threshold drives
-
Drive systems wherein the release of only a few individuals is required for super-Mendelian spread of the drive.
- Essential gene
-
A gene required for the viability or reproduction of an organism.
- Homology-directed repair
-
(HDR). A DNA repair pathway initiated following induction of double-strand DNA breaks in which the break is repaired by copying sequences (or a template) from an identical sister chromosome (typically following DNA replication in somatic cells) or from the homologous chromosome (typically during meiosis, although sometimes also in somatic cells).
- Anopheline mosquitoes
-
A genus of mosquitoes that carry malarial parasites.
- CRISPR
-
A bacterial immunity system from which the synthetic CRISPR–Cas9 genome editing system was derived by Jenifer Doudna and colleagues.
- Guide RNA
-
(gRNA). A synthetic linkage of two RNA components of bacterial CRISPR systems: transactivating CRISPR RNA and CRISPR RNA. gRNAs are sometimes also denoted sgRNA (synthetic-guide RNA or single-guide RNA).
- Anti-malarial effectors
-
Transgenes encoding proteins that, when expressed, exert a desired anti-malarial effect, such as for anti-malarial peptides expressed following a blood meal in mosquitoes.
- Trans-complementing drive
-
A split CRISPR-based gene-drive system comprising two components: a Cas9-expressing element and a guide RNA (gRNA)-bearing element that are inserted at different sites in the genome. The gRNA element carries two gRNAs, one to copy itself and the other cutting at the insertion site of the Cas9 element. Alone, each element is inherited in a standard Mendelian fashion. When combined, however, the gRNAs complex with Cas9 provided in trans to drive copying of both elements, thereby creating a full-drive system that efficiently transmits both elements to progeny.
- Neutralizing drives
-
Genetic elements designed to eliminate or halt the spread of a gene drive. Examples include active guide RNA (gRNA)-only elements such as e-CHACRs (erasing constructs hitchhiking on the autocatalytic chain reaction), ERACRs (elements reversing the autocatalytic chain reaction) or Cas9-triggered chain ablation (CATCHA) elements, anti-Cas9 proteins, as well as passive recoded fitness-neutral gRNA target sites (for replacing suppression drives).
- Active genetics
-
Copying of a genetic element from one chromosome to its homologue in response to a double-strand DNA break being generated in the homologue at the same genomic site where the active genetic element is inserted. Copying, which results from directional gene conversion, is typically mediated in the germline by the synthesis-dependent strand annealing (or D-loop) branch of the homology-directed repair pathway.
- Suppression drives
-
Gene drives that reduce the size of the target population. Typically, suppression drives insert into genes required for the viability or fertility of one or both sexes.
- Malaria
-
An infectious blood disease caused by Plasmodial parasites (for example, Plasmodium falciparum and Plasmodium vivax) that is primarily transmitted (vectored) by mosquitoes in the Anopheline genus.
- Aedes mosquitoes
-
A genus of mosquitoes carrying arboviruses that transmit dengue fever, yellow fever, chikungunya or Zika.
- Modification drives
-
Also referred to as replacement drives. Drive systems that modify a target population but do not reduce its numbers.
- Non-homologous end-joining
-
(NHEJ). A DNA repair pathway initiated following the induction of double-strand DNA breaks in which the two ends of DNA are ligated back to each other in a template-independent fashion. NHEJ can introduce small insertions or deletions, which can cause mutations and frameshift-based loss-of-function alleles of a gene.
- Basic reproductive number
-
(R0). The expected number of disease cases generated by one person infected with a pathogen if all individuals are susceptible to infection (that is, at the beginning of disease spread). For vector-borne diseases, it depends on several factors involving the vector hosts as well as infected persons (Box 1).
- Introduction
-
The full introduction of a specific allele from one genetic background into another results in all progeny carrying the allele in question. The term introgression, typically of a trait that is potentially associated with additional surrounding local genetic variation from one strain into another background, can also be used in such contexts. Example 1: a gene-drive element carried in one strain that spreads to 100% introduction throughout a naive target population. Example 2: an insecticide-resistant allele and a neighbouring 1 megabase region were introgressed into a sensitive strain of mosquitoes from a resistant strain.
- Recoded drives
-
Gene drives that carry a recoded version of a target gene into which it inserts such that the recoded sequences at the 5´ end of the drive element are fused seamlessly to endogenous coding sequences, restoring wild-type protein activity and expression of the endogenous gene.
- CHACRs
-
(Constructs hitchhiking on the autocatalytic chain reaction). Active genetic elements carrying a guide RNA to copy themselves and potential cargo but no Cas9 source.
- Allelic drives
-
Genetic systems biasing the inheritance of a particular allelic variant, typically altering only one or a few base pairs. CRISPR-based allelic-drive systems can be of two types, copy-cutting or copy-grafting.
- e-CHACR
-
(erasing construct hitchhiking on the autocatalytic chain reaction). A drive-neutralizing active genetic element that encodes two or more guide RNAs (gRNAs) but no source of Cas9. One of the gRNAs cuts at the genomic site of e-CHACR insertion, enabling self-copying in the presence of a trans-acting source of Cas9 provided by the gene drive. The additional gRNAs target cleavage and inactivation of the Cas9 transgene component of a gene-drive element. e-CHACRs can be inserted into the genome at any desired location.
- Copy-cutting
-
A form of allelic drive in which the driving guide RNA directly cuts the non-preferred allelic variant, leaving the preferred allele intact.
- Copy-grafting
-
A form of allelic drive in which the preferred allele is adjacent to a sequence immune to cleavage by a guide RNA (gRNA) that can cut wild-type or non-preferred alleles. When the gRNA cuts either the wild-type or non-preferred allelic variants, the double-strand break is repaired by homology-directed repair (HDR) using the cleavage-resistant allele as a template, copying the favoured allelic variant in the process. The cleavage-resistant gRNA site and preferred allele are typically <25 bp apart to be copied reliably together as a by-product of the short-range end-resection step of HDR.
- ERACRs
-
(Element reversing the autocatalytic chain reaction). Drive-neutralizing active genetic elements that encode two guide RNAs cutting on either side of the gene-drive element. ERACRs are inserted at the same genomic site as the drive elements they are designed to delete and replace. ERACRs can carry additional cargo such as recoded copies of a gene targeted for disruption by the gene drive. However, ERACRs do not carry a Cas9 source.
- Allo-alleles
-
Alleles in a polyploid species (allopolyploid or autoploid) whose chromosomes either derive from whole-genome duplications within a species (autoploidy) or result from the fusion of two different genomes (alloploidy) followed by the potential partial loss of genetic information. Many crop species such as grains (wheat, rye), corn, peanuts and sugarcane are polyploid as are a few animal species (for example, Xenopus laevis (African clawed frogs)).
- Culex mosquitoes
-
A genus of mosquitoes serving as vectors for diseases, including West Nile fever, St Louis encephalitis, Japanese encephalitis, and viral diseases in birds and horses.
- World Health Organization phases of gene-drive development
-
The phases are as follows. Phase 1 is laboratory development and testing. Phase 2 is confined outdoor tests (either in large cages or isolated environments such as islands); the goal of these experiments is to achieve defined entomological endpoints such as a certain percentage of population reduction (suppression) or drive introgression (modification). Phase 3 is limited testing in disease-endemic regions to demonstrate epidemiological efficacy. Phase 4 is full implementation.
- Risk assessment
-
A formal analysis of all factors that need to be considered, evaluated and balanced before implementing an intervention such as the release of organisms carrying a gene-drive element. Risks should include predicted or potential consequences associated with performing the intervention as well as those accompanying a decision not to implement the technology.
Rights and permissions
About this article
Cite this article
Bier, E. Gene drives gaining speed. Nat Rev Genet 23, 5–22 (2022). https://doi.org/10.1038/s41576-021-00386-0
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41576-021-00386-0
This article is cited by
-
Generating prophylactic immunity against arboviruses in vertebrates and invertebrates
Nature Reviews Immunology (2024)
-
Genetic and geographic population structure in the malaria vector, Anopheles farauti, provides a candidate system for pioneering confinable gene-drive releases
Heredity (2024)
-
CRISPR–Cas system to discover host-virus interactions in Flaviviridae
Virology Journal (2023)
-
Advances in the genetic characterization of the malaria vector, Anopheles funestus, and implications for improved surveillance and control
Malaria Journal (2023)
-
Enhancing the scalability of Wolbachia-based vector-borne disease management: time and temperature limits for storage and transport of Wolbachia-infected Aedes aegypti eggs for field releases
Parasites & Vectors (2023)